Difference between revisions of "Matplotlib DPL94 R1rho R2eff"
(→Code) |
|||
| Line 71: | Line 71: | ||
import sys | import sys | ||
import os | import os | ||
| − | from math import cos, sin | + | from math import cos, sin, sqrt, pi |
from numpy import array, float64 | from numpy import array, float64 | ||
| − | + | ||
### plotting facility. | ### plotting facility. | ||
import matplotlib.pyplot as plt | import matplotlib.pyplot as plt | ||
| − | + | ||
# Ordered dictionary | # Ordered dictionary | ||
import collections | import collections | ||
| − | + | ||
### relax modules | ### relax modules | ||
# Import some tools to loop over the spins. | # Import some tools to loop over the spins. | ||
| Line 86: | Line 86: | ||
from specific_analyses.relax_disp.disp_data import calc_rotating_frame_params, generate_r20_key, loop_exp_frq, loop_exp_frq_offset, loop_point, return_param_key_from_data, return_spin_lock_nu1 | from specific_analyses.relax_disp.disp_data import calc_rotating_frame_params, generate_r20_key, loop_exp_frq, loop_exp_frq_offset, loop_point, return_param_key_from_data, return_spin_lock_nu1 | ||
from specific_analyses.relax_disp import optimisation | from specific_analyses.relax_disp import optimisation | ||
| − | + | from lib.nmr import frequency_to_Hz, frequency_to_ppm, frequency_to_rad_per_s | |
| + | |||
############### | ############### | ||
| − | + | ||
# You have to provide a DPL94 results state file | # You have to provide a DPL94 results state file | ||
res_folder = "resultsR1" | res_folder = "resultsR1" | ||
#res_folder = "results_clustering" | #res_folder = "results_clustering" | ||
res_state = os.path.join(res_folder, "DPL94", "results") | res_state = os.path.join(res_folder, "DPL94", "results") | ||
| − | spin_inte = ": | + | spin_inte = ":44@N" |
| − | + | # Make a fake spin, from the spin of interest | |
| + | fake_spin_inte = spin_inte.replace("N","X") | ||
| + | |||
# Interpolate graph settings | # Interpolate graph settings | ||
#num_points=1000, extend=500.0 | #num_points=1000, extend=500.0 | ||
num_points=100 | num_points=100 | ||
extend=5000.0 | extend=5000.0 | ||
| − | + | ||
################ | ################ | ||
spin_inte_rep = spin_inte.replace('#', '_').replace(':', '_').replace('@', '_') | spin_inte_rep = spin_inte.replace('#', '_').replace(':', '_').replace('@', '_') | ||
| − | + | ||
# Load the state | # Load the state | ||
state.load(res_state, force=True) | state.load(res_state, force=True) | ||
| + | # Get the dictionary key | ||
| + | for exp_type, frq in loop_exp_frq(): | ||
| + | r20_key = generate_r20_key(exp_type=exp_type, frq=frq) | ||
| + | |||
# Show pipes | # Show pipes | ||
pipe.display() | pipe.display() | ||
pipe.current() | pipe.current() | ||
| − | + | ||
# Get the spin of interest and save it in cdp, to access it after execution of script. | # Get the spin of interest and save it in cdp, to access it after execution of script. | ||
cdp.myspin = return_spin(spin_inte) | cdp.myspin = return_spin(spin_inte) | ||
| + | |||
| + | # Copy the parameters from spin of interest to a fake spin to be modified. | ||
| + | spin.copy(spin_from=spin_inte, spin_to=fake_spin_inte) | ||
| + | # Returnspin | ||
| + | cdp.fakespin = return_spin(fake_spin_inte) | ||
| + | |||
| + | # Modify data | ||
| + | if spin_inte == ":52@N": | ||
| + | # Set reference data | ||
| + | cdp.fakespin.r2[r20_key] = 6.51945 | ||
| + | cdp.fakespin.kex = 13193.82986 | ||
| + | cdp.fakespin.kex_err = 2307.09152 | ||
| + | phi_ex_rad2_s2 = 93499.92172 | ||
| + | phi_ex_err_rad2_s2 = 33233.23039 | ||
| + | scaling_rad2_s2 = frequency_to_ppm(frq=1/(2*pi), B0=cdp.spectrometer_frq_list[0], isotope='15N')**2 | ||
| + | print scaling_rad2_s2 | ||
| + | |||
| + | cdp.fakespin.phi_ex = phi_ex_rad2_s2*scaling_rad2_s2 | ||
| + | cdp.fakespin.phi_ex_err = phi_ex_err_rad2_s2*scaling_rad2_s2 | ||
| + | |||
| + | print cdp.myspin.ri_data['R1'], cdp.myspin.ri_data_err['R1'], cdp.myspin.r2[r20_key], cdp.myspin.kex, cdp.myspin.phi_ex | ||
| + | print cdp.fakespin.ri_data['R1'], cdp.fakespin.ri_data_err['R1'], cdp.fakespin.r2[r20_key], cdp.fakespin.kex, cdp.fakespin.phi_ex | ||
| + | |||
# Calculate the offset data | # Calculate the offset data | ||
| − | theta_spin_dic, Domega_spin_dic, w_eff_spin_dic, dic_key_list = calc_rotating_frame_params(spin=cdp.myspin, spin_id=spin_inte, verbosity= | + | theta_spin_dic, Domega_spin_dic, w_eff_spin_dic, dic_key_list = calc_rotating_frame_params(spin=cdp.myspin, spin_id=spin_inte, verbosity=0) |
# Save the data in cdp to access it after execution of script. | # Save the data in cdp to access it after execution of script. | ||
cdp.myspin.theta_spin_dic = theta_spin_dic | cdp.myspin.theta_spin_dic = theta_spin_dic | ||
cdp.myspin.w_eff_spin_dic = w_eff_spin_dic | cdp.myspin.w_eff_spin_dic = w_eff_spin_dic | ||
cdp.myspin.dic_key_list = dic_key_list | cdp.myspin.dic_key_list = dic_key_list | ||
| − | + | ||
############################ | ############################ | ||
# First creacte back calculated R2eff data for interpolated plots. | # First creacte back calculated R2eff data for interpolated plots. | ||
############################ | ############################ | ||
| + | |||
| + | |||
| + | # Return the original structure for frq, offset | ||
| + | spin_lock_nu1 = return_spin_lock_nu1(ref_flag=False) | ||
| + | # Back calculate R2eff data for the set parameters. | ||
| + | cdp.fakespin.back_calc = optimisation.back_calc_r2eff(spin=cdp.fakespin, spin_id=fake_spin_inte, spin_lock_nu1=spin_lock_nu1) | ||
| − | |||
| − | |||
# Prepare list to hold new data | # Prepare list to hold new data | ||
spin_lock_nu1_new = [] | spin_lock_nu1_new = [] | ||
| − | |||
# Loop over the structures to generate data | # Loop over the structures to generate data | ||
for ei in range(len(spin_lock_nu1)): | for ei in range(len(spin_lock_nu1)): | ||
# Add a new dimension. | # Add a new dimension. | ||
spin_lock_nu1_new.append([]) | spin_lock_nu1_new.append([]) | ||
| − | + | ||
# Then loop over the spectrometer frequencies. | # Then loop over the spectrometer frequencies. | ||
for mi in range(len(spin_lock_nu1[ei])): | for mi in range(len(spin_lock_nu1[ei])): | ||
# Add a new dimension. | # Add a new dimension. | ||
spin_lock_nu1_new[ei].append([]) | spin_lock_nu1_new[ei].append([]) | ||
| − | + | ||
# Finally the offsets. | # Finally the offsets. | ||
for oi in range(len(spin_lock_nu1[ei][mi])): | for oi in range(len(spin_lock_nu1[ei][mi])): | ||
# Add a new dimension. | # Add a new dimension. | ||
spin_lock_nu1_new[ei][mi].append([]) | spin_lock_nu1_new[ei][mi].append([]) | ||
| − | + | ||
# No data. | # No data. | ||
if not len(spin_lock_nu1[ei][mi][oi]): | if not len(spin_lock_nu1[ei][mi][oi]): | ||
continue | continue | ||
| − | + | ||
# Interpolate (adding the extended amount to the end). | # Interpolate (adding the extended amount to the end). | ||
for di in range(num_points): | for di in range(num_points): | ||
| Line 155: | Line 188: | ||
# Intersert field 0 | # Intersert field 0 | ||
#spin_lock_nu1_new[ei][mi][oi][0] = 0.0 | #spin_lock_nu1_new[ei][mi][oi][0] = 0.0 | ||
| − | + | ||
# Convert to a numpy array. | # Convert to a numpy array. | ||
spin_lock_nu1_new[ei][mi][oi] = array(spin_lock_nu1_new[ei][mi][oi], float64) | spin_lock_nu1_new[ei][mi][oi] = array(spin_lock_nu1_new[ei][mi][oi], float64) | ||
| − | + | ||
# Then back calculate R2eff data for the interpolated points. | # Then back calculate R2eff data for the interpolated points. | ||
cdp.myspin.back_calc = optimisation.back_calc_r2eff(spin=cdp.myspin, spin_id=spin_inte, spin_lock_nu1=spin_lock_nu1_new) | cdp.myspin.back_calc = optimisation.back_calc_r2eff(spin=cdp.myspin, spin_id=spin_inte, spin_lock_nu1=spin_lock_nu1_new) | ||
| − | + | ||
# Calculate the offset data, interpolated | # Calculate the offset data, interpolated | ||
theta_spin_dic_inter, Domega_spin_dic_inter, w_eff_spin_dic_inter, dic_key_list_inter = calc_rotating_frame_params(spin=cdp.myspin, spin_id=spin_inte, fields = spin_lock_nu1_new, verbosity=0) | theta_spin_dic_inter, Domega_spin_dic_inter, w_eff_spin_dic_inter, dic_key_list_inter = calc_rotating_frame_params(spin=cdp.myspin, spin_id=spin_inte, fields = spin_lock_nu1_new, verbosity=0) | ||
| − | + | ||
###### Store the data before plotting | ###### Store the data before plotting | ||
# Create a dictionary to hold data | # Create a dictionary to hold data | ||
cdp.mydic = collections.OrderedDict() | cdp.mydic = collections.OrderedDict() | ||
| − | + | ||
# Loop over the data structures and save to dictionary | # Loop over the data structures and save to dictionary | ||
for exp_type, frq, offset, ei, mi, oi in loop_exp_frq_offset(return_indices=True): | for exp_type, frq, offset, ei, mi, oi in loop_exp_frq_offset(return_indices=True): | ||
| − | |||
| − | |||
# This is not used, but could be used to get Rex. | # This is not used, but could be used to get Rex. | ||
R1_rho_prime = cdp.myspin.r2[r20_key] | R1_rho_prime = cdp.myspin.r2[r20_key] | ||
| − | + | #print R1_rho_prime | |
| + | |||
# Get R1 | # Get R1 | ||
R1 = cdp.myspin.ri_data['R1'] | R1 = cdp.myspin.ri_data['R1'] | ||
R1_err = cdp.myspin.ri_data_err['R1'] | R1_err = cdp.myspin.ri_data_err['R1'] | ||
| − | + | ||
# Add to dic | # Add to dic | ||
if exp_type not in cdp.mydic: | if exp_type not in cdp.mydic: | ||
| Line 199: | Line 231: | ||
cdp.mydic[exp_type][frq][offset]['R1_rho_bc'] = [] | cdp.mydic[exp_type][frq][offset]['R1_rho_bc'] = [] | ||
cdp.mydic[exp_type][frq][offset]['R1_rho_inter'] = [] | cdp.mydic[exp_type][frq][offset]['R1_rho_inter'] = [] | ||
| + | |||
| + | # Y val fake | ||
| + | cdp.mydic[exp_type][frq][offset]['fake_R1_rho'] = [] | ||
| + | |||
# Y2 val | # Y2 val | ||
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff'] = [] | cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff'] = [] | ||
| Line 204: | Line 240: | ||
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_bc'] = [] | cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_bc'] = [] | ||
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_inter'] = [] | cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_inter'] = [] | ||
| − | + | ||
# Loop over the original dispersion points. | # Loop over the original dispersion points. | ||
for point, di in loop_point(exp_type=exp_type, frq=frq, offset=offset, return_indices=True): | for point, di in loop_point(exp_type=exp_type, frq=frq, offset=offset, return_indices=True): | ||
param_key = return_param_key_from_data(exp_type=exp_type, frq=frq, offset=offset, point=point) | param_key = return_param_key_from_data(exp_type=exp_type, frq=frq, offset=offset, point=point) | ||
| − | + | ||
# X val | # X val | ||
cdp.mydic[exp_type][frq][offset]['point'].append(point) | cdp.mydic[exp_type][frq][offset]['point'].append(point) | ||
| Line 215: | Line 251: | ||
w_eff = w_eff_spin_dic[param_key] | w_eff = w_eff_spin_dic[param_key] | ||
cdp.mydic[exp_type][frq][offset]['w_eff'].append(w_eff) | cdp.mydic[exp_type][frq][offset]['w_eff'].append(w_eff) | ||
| − | + | ||
# Average resonance spin_lock_offset | # Average resonance spin_lock_offset | ||
#print Domega_spin_dic[param_key] | #print Domega_spin_dic[param_key] | ||
| − | + | ||
# Y val | # Y val | ||
R1_rho = cdp.myspin.r2eff[param_key] | R1_rho = cdp.myspin.r2eff[param_key] | ||
| Line 226: | Line 262: | ||
R1_rho_bc = cdp.myspin.r2eff_bc[param_key] | R1_rho_bc = cdp.myspin.r2eff_bc[param_key] | ||
cdp.mydic[exp_type][frq][offset]['R1_rho_bc'].append(R1_rho_bc) | cdp.mydic[exp_type][frq][offset]['R1_rho_bc'].append(R1_rho_bc) | ||
| + | |||
| + | # Y val, fake | ||
| + | fake_R1_rho = cdp.fakespin.back_calc[ei][0][mi][oi][di] | ||
| + | cdp.mydic[exp_type][frq][offset]['fake_R1_rho'].append(fake_R1_rho) | ||
# Y2 val | # Y2 val | ||
| Line 231: | Line 271: | ||
R1_rho_R2eff = (R1_rho - R1*cos(theta)*cos(theta)) / (sin(theta) * sin(theta)) | R1_rho_R2eff = (R1_rho - R1*cos(theta)*cos(theta)) / (sin(theta) * sin(theta)) | ||
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff'].append(R1_rho_R2eff) | cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff'].append(R1_rho_R2eff) | ||
| − | + | ||
R1_rho_R2eff_err = (R1_rho_err - R1_err*cos(theta)*cos(theta)) / (sin(theta) * sin(theta)) | R1_rho_R2eff_err = (R1_rho_err - R1_err*cos(theta)*cos(theta)) / (sin(theta) * sin(theta)) | ||
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_err'].append(R1_rho_R2eff_err) | cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_err'].append(R1_rho_R2eff_err) | ||
| − | + | ||
R1_rho_R2eff_bc = (R1_rho_bc - R1*cos(theta)*cos(theta)) / (sin(theta) * sin(theta)) | R1_rho_R2eff_bc = (R1_rho_bc - R1*cos(theta)*cos(theta)) / (sin(theta) * sin(theta)) | ||
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_bc'].append(R1_rho_R2eff_bc) | cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_bc'].append(R1_rho_R2eff_bc) | ||
| − | + | ||
## Loop over the new dispersion points. | ## Loop over the new dispersion points. | ||
for di in range(len(cdp.myspin.back_calc[ei][0][mi][oi])): | for di in range(len(cdp.myspin.back_calc[ei][0][mi][oi])): | ||
point = spin_lock_nu1_new[ei][mi][oi][di] | point = spin_lock_nu1_new[ei][mi][oi][di] | ||
param_key = return_param_key_from_data(exp_type=exp_type, frq=frq, offset=offset, point=point) | param_key = return_param_key_from_data(exp_type=exp_type, frq=frq, offset=offset, point=point) | ||
| − | + | ||
# X val | # X val | ||
cdp.mydic[exp_type][frq][offset]['point_inter'].append(point) | cdp.mydic[exp_type][frq][offset]['point_inter'].append(point) | ||
| Line 249: | Line 289: | ||
w_eff = w_eff_spin_dic_inter[param_key] | w_eff = w_eff_spin_dic_inter[param_key] | ||
cdp.mydic[exp_type][frq][offset]['w_eff_inter'].append(w_eff) | cdp.mydic[exp_type][frq][offset]['w_eff_inter'].append(w_eff) | ||
| − | + | ||
# Y val | # Y val | ||
R1_rho = cdp.myspin.back_calc[ei][0][mi][oi][di] | R1_rho = cdp.myspin.back_calc[ei][0][mi][oi][di] | ||
cdp.mydic[exp_type][frq][offset]['R1_rho_inter'].append(R1_rho) | cdp.mydic[exp_type][frq][offset]['R1_rho_inter'].append(R1_rho) | ||
| − | + | ||
# Y2 val | # Y2 val | ||
# Calc R1_rho_R2eff | # Calc R1_rho_R2eff | ||
R1_rho_R2eff = (R1_rho - R1*cos(theta)*cos(theta)) / (sin(theta) * sin(theta)) | R1_rho_R2eff = (R1_rho - R1*cos(theta)*cos(theta)) / (sin(theta) * sin(theta)) | ||
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_inter'].append(R1_rho_R2eff) | cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_inter'].append(R1_rho_R2eff) | ||
| − | + | ||
#if oi == 0: | #if oi == 0: | ||
#print exp_type, frq, offset, point, theta, w_eff | #print exp_type, frq, offset, point, theta, w_eff | ||
| − | + | ||
####### PLOT #### | ####### PLOT #### | ||
| − | + | ||
## Define labels for plotting | ## Define labels for plotting | ||
filesave_R1_rho_R2eff = 'R1_rho_R2eff' | filesave_R1_rho_R2eff = 'R1_rho_R2eff' | ||
filesave_R1_rho = 'R1_rho' | filesave_R1_rho = 'R1_rho' | ||
| − | + | ||
# For writing math in matplotlib, see | # For writing math in matplotlib, see | ||
# http://matplotlib.org/1.3.1/users/mathtext.html | # http://matplotlib.org/1.3.1/users/mathtext.html | ||
| − | + | ||
ylabel_R1_rho = r'R$_{1\rho}$ [rad s$^{-1}$]' | ylabel_R1_rho = r'R$_{1\rho}$ [rad s$^{-1}$]' | ||
ylabel_R1_rho_R2eff = r'R$_{1\rho, R_{2,eff}}$ [rad s$^{-1}$]' | ylabel_R1_rho_R2eff = r'R$_{1\rho, R_{2,eff}}$ [rad s$^{-1}$]' | ||
| − | + | ||
xlabel_theta = 'Rotating frame tilt angle [rad]' | xlabel_theta = 'Rotating frame tilt angle [rad]' | ||
xlabel_w_eff = r'Effective field in rotating frame [rad s$^{-1}$]' | xlabel_w_eff = r'Effective field in rotating frame [rad s$^{-1}$]' | ||
xlabel_lock = 'Spin-lock field strength [Hz]' | xlabel_lock = 'Spin-lock field strength [Hz]' | ||
| − | + | ||
# Set image inches size | # Set image inches size | ||
img_inch_x = 12 | img_inch_x = 12 | ||
img_inch_y = img_inch_x / 1.6 | img_inch_y = img_inch_x / 1.6 | ||
legend_size = 6 | legend_size = 6 | ||
| − | + | ||
| − | + | ||
# Plot values in dic | # Plot values in dic | ||
for exptype, frq_dic in cdp.mydic.items(): | for exptype, frq_dic in cdp.mydic.items(): | ||
| Line 292: | Line 332: | ||
graphlabel_bc = "%3.1f_%3.3f_bc"%(frq/1E6, offset) | graphlabel_bc = "%3.1f_%3.3f_bc"%(frq/1E6, offset) | ||
graphlabel_inter = "%3.1f_%3.3f_inter"%(frq/1E6, offset) | graphlabel_inter = "%3.1f_%3.3f_inter"%(frq/1E6, offset) | ||
| − | + | graphlabel_fake = "%3.1f_%3.3f_fake"%(frq/1E6, offset) | |
| + | |||
# Plot 1: R1_rho as function of theta. | # Plot 1: R1_rho as function of theta. | ||
plt.figure(1) | plt.figure(1) | ||
| Line 298: | Line 339: | ||
plt.errorbar(val_dics['theta'], val_dics['R1_rho'], yerr=val_dics['R1_rho_err'], fmt='o', label=graphlabel, color=line.get_color()) | plt.errorbar(val_dics['theta'], val_dics['R1_rho'], yerr=val_dics['R1_rho_err'], fmt='o', label=graphlabel, color=line.get_color()) | ||
plt.plot(val_dics['theta'], val_dics['R1_rho_bc'], 'D', label=graphlabel_bc, color=line.get_color()) | plt.plot(val_dics['theta'], val_dics['R1_rho_bc'], 'D', label=graphlabel_bc, color=line.get_color()) | ||
| − | + | ||
# Plot 2: R1_rho_R2eff as function of w_eff | # Plot 2: R1_rho_R2eff as function of w_eff | ||
plt.figure(2) | plt.figure(2) | ||
| Line 309: | Line 350: | ||
plt.errorbar(val_dics['w_eff'], val_dics['R1_rho_R2eff'], yerr=val_dics['R1_rho_R2eff_err'], fmt='o', label=graphlabel, color=line.get_color()) | plt.errorbar(val_dics['w_eff'], val_dics['R1_rho_R2eff'], yerr=val_dics['R1_rho_R2eff_err'], fmt='o', label=graphlabel, color=line.get_color()) | ||
plt.plot(val_dics['w_eff'], val_dics['R1_rho_R2eff_bc'], 'D', label=graphlabel_bc, color=line.get_color()) | plt.plot(val_dics['w_eff'], val_dics['R1_rho_R2eff_bc'], 'D', label=graphlabel_bc, color=line.get_color()) | ||
| − | + | ||
# Plot 3: R1_rho as function of as function of disp_point, the Spin-lock field strength | # Plot 3: R1_rho as function of as function of disp_point, the Spin-lock field strength | ||
plt.figure(3) | plt.figure(3) | ||
| Line 315: | Line 356: | ||
plt.errorbar(val_dics['point'], val_dics['R1_rho'], yerr=val_dics['R1_rho_err'], fmt='o', label=graphlabel, color=line.get_color()) | plt.errorbar(val_dics['point'], val_dics['R1_rho'], yerr=val_dics['R1_rho_err'], fmt='o', label=graphlabel, color=line.get_color()) | ||
plt.plot(val_dics['point'], val_dics['R1_rho_bc'], 'D', label=graphlabel_bc, color=line.get_color()) | plt.plot(val_dics['point'], val_dics['R1_rho_bc'], 'D', label=graphlabel_bc, color=line.get_color()) | ||
| − | + | plt.plot(val_dics['point'], val_dics['fake_R1_rho'], '*', label=graphlabel_fake, color=line.get_color()) | |
| − | + | ||
| + | |||
# Define settings for each graph | # Define settings for each graph | ||
# Plot 1: R1_rho as function of theta. | # Plot 1: R1_rho as function of theta. | ||
| Line 324: | Line 366: | ||
plt.legend(loc='best', prop={'size':legend_size}) | plt.legend(loc='best', prop={'size':legend_size}) | ||
plt.grid(True) | plt.grid(True) | ||
| − | plt.ylim([0,16]) | + | #plt.ylim([0,16]) |
plt.title("%s \n %s as function of %s"%(spin_inte, ylabel_R1_rho, xlabel_theta)) | plt.title("%s \n %s as function of %s"%(spin_inte, ylabel_R1_rho, xlabel_theta)) | ||
fig1.set_size_inches(img_inch_x, img_inch_y) | fig1.set_size_inches(img_inch_x, img_inch_y) | ||
plt.savefig("matplotlib_%s_%s_theta_sep.png"%(spin_inte_rep, filesave_R1_rho) ) | plt.savefig("matplotlib_%s_%s_theta_sep.png"%(spin_inte_rep, filesave_R1_rho) ) | ||
| − | + | ||
## Plot 2: R1_rho_R2eff as function of w_eff | ## Plot 2: R1_rho_R2eff as function of w_eff | ||
fig2 = plt.figure(2) | fig2 = plt.figure(2) | ||
| Line 335: | Line 377: | ||
plt.legend(loc='best', prop={'size':legend_size}) | plt.legend(loc='best', prop={'size':legend_size}) | ||
plt.grid(True) | plt.grid(True) | ||
| − | plt.ylim([0,16]) | + | #plt.ylim([0,16]) |
#plt.xlim([0,20000*20000]) | #plt.xlim([0,20000*20000]) | ||
plt.xlim([0,20000]) | plt.xlim([0,20000]) | ||
| Line 341: | Line 383: | ||
fig2.set_size_inches(img_inch_x, img_inch_y) | fig2.set_size_inches(img_inch_x, img_inch_y) | ||
plt.savefig("matplotlib_%s_%s_w_eff.png"%(spin_inte_rep, filesave_R1_rho_R2eff) ) | plt.savefig("matplotlib_%s_%s_w_eff.png"%(spin_inte_rep, filesave_R1_rho_R2eff) ) | ||
| − | + | ||
## Plot 3: R1_rho as function of as function of disp_point, the Spin-lock field strength | ## Plot 3: R1_rho as function of as function of disp_point, the Spin-lock field strength | ||
fig3 = plt.figure(3) | fig3 = plt.figure(3) | ||
| Line 348: | Line 390: | ||
plt.legend(loc='best', prop={'size':legend_size}) | plt.legend(loc='best', prop={'size':legend_size}) | ||
plt.grid(True) | plt.grid(True) | ||
| − | plt.ylim([0,16]) | + | #plt.ylim([0,16]) |
plt.title("%s \n %s as function of %s"%(spin_inte, ylabel_R1_rho, xlabel_lock)) | plt.title("%s \n %s as function of %s"%(spin_inte, ylabel_R1_rho, xlabel_lock)) | ||
fig3.set_size_inches(img_inch_x, img_inch_y) | fig3.set_size_inches(img_inch_x, img_inch_y) | ||
Revision as of 15:33, 18 March 2014
Contents
About
The production to these figures relates to the Suppport Request:
sr #3124: Grace graphs production for R1rho analysis with R2_eff as function of Omega_eff
References
Refer to the manual for parameter explanation
- Evenäs, J., Malmendal, A. & Akke, M. (2001). Dynamics of the transition between open and closed conformations in a calmodulin C-terminal domain mutant. Structure 9, 185–195 DOI
- Kempf, J.G. & Loria, J.P. (2004). Measurement of intermediate exchange phenomena. Methods Mol. Biol. 278, 185–231 DOI
- Palmer, A.G. & Massi, F. (2006). Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem. Rev. 106, 1700–1719 DOI
- Palmer, A.G., Kroenke, C.D. & Loria, J.P. (2001). Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Meth. Enzymol. 339 DOI
- Francesca Massi, Michael J. Grey, Arthur G. Palmer III* (2005) Microsecond timescale backbone conformational dynamics in ubiquitin studied with NMR R1ρ relaxation experiments, Protein science DOI
Figures
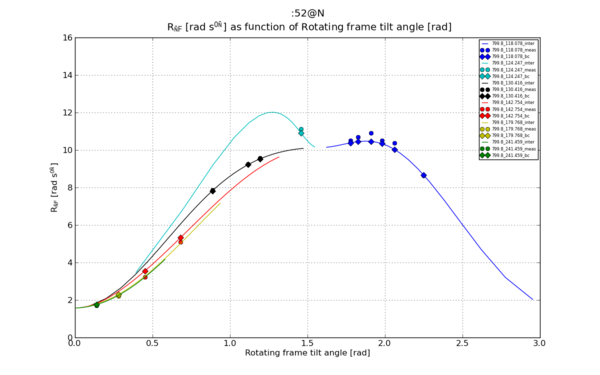
Ref [1], Figure 1.b. The bell-curves As function of angle calculation.
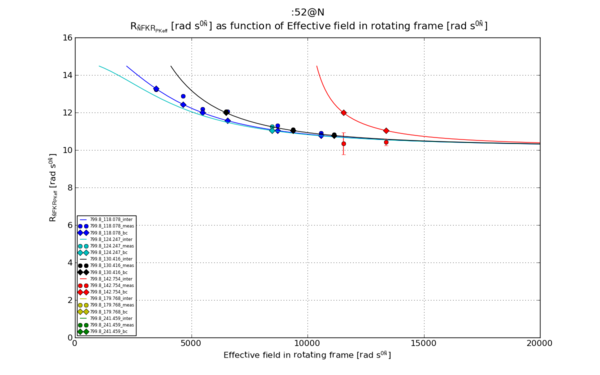
Ref [1], Figure 1.c. The wanted graph. No clear "name" for the calculated parameter.
Ref [2], Equation 27.
Here the calculated value is noted as: $R_{eff}$. : Equation 27: $ R_{eff} = R_{1\rho} / sin^2(\theta) - R_1 / tan^2(\theta) = R^{0}_2 + R_{ex} $.
Where $R^{0}_2$ refers to $R_{1\rho '}$ as seen at DPL94
Ref [3], Equation 20.
Here the calculated value is noted as: $R_2$: $R_2 = R_{1\rho} / sin^2(\theta) - R_1 / tan^2(\theta)$
Figure 11+16, would be the reference.
Ref [4], Equation 43. $R_{eff} = R_{1\rho} / sin^2(\theta) - R_1 / tan^2(\theta)$
Ref [5], Material and Methods, page 740. Here the calculated value is noted as: $R_2: R_2 = R^{0}_2 + R_{ex}$.
Figure 4 would be the wished graphs.
A little table of conversion then gives
Relax equation | Relax store | Articles
---------------------------------------------------------------
R1rho' spin.r2 R^{0}_2 or Bar{R}_2
Fitted pars Not stored R_ex
R1rho spin.r2eff R1rho
R_1 spin.ri_data['R1'] R_1 or Bar{R}_1
The parameter is called R_2 or R_eff in the articles. Since reff is not used in relax, this could be used?
A description could be:
- The effective rate
- The effective transverse relaxation rate constant
- The effective relaxation rate constant.
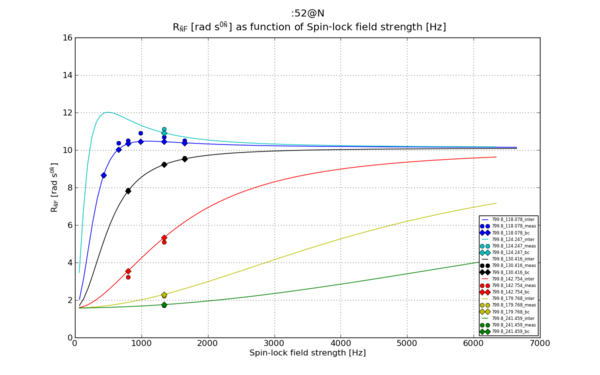
Make graphs
The outcome
To run
relax -p r1rhor2eff.py
Code
File: r1rhor2eff.py
### python imports
import sys
import os
from math import cos, sin, sqrt, pi
from numpy import array, float64
### plotting facility.
import matplotlib.pyplot as plt
# Ordered dictionary
import collections
### relax modules
# Import some tools to loop over the spins.
from pipe_control.mol_res_spin import return_spin, spin_loop
# Import method to calculate the R1_rho offset data
from specific_analyses.relax_disp.disp_data import calc_rotating_frame_params, generate_r20_key, loop_exp_frq, loop_exp_frq_offset, loop_point, return_param_key_from_data, return_spin_lock_nu1
from specific_analyses.relax_disp import optimisation
from lib.nmr import frequency_to_Hz, frequency_to_ppm, frequency_to_rad_per_s
###############
# You have to provide a DPL94 results state file
res_folder = "resultsR1"
#res_folder = "results_clustering"
res_state = os.path.join(res_folder, "DPL94", "results")
spin_inte = ":44@N"
# Make a fake spin, from the spin of interest
fake_spin_inte = spin_inte.replace("N","X")
# Interpolate graph settings
#num_points=1000, extend=500.0
num_points=100
extend=5000.0
################
spin_inte_rep = spin_inte.replace('#', '_').replace(':', '_').replace('@', '_')
# Load the state
state.load(res_state, force=True)
# Get the dictionary key
for exp_type, frq in loop_exp_frq():
r20_key = generate_r20_key(exp_type=exp_type, frq=frq)
# Show pipes
pipe.display()
pipe.current()
# Get the spin of interest and save it in cdp, to access it after execution of script.
cdp.myspin = return_spin(spin_inte)
# Copy the parameters from spin of interest to a fake spin to be modified.
spin.copy(spin_from=spin_inte, spin_to=fake_spin_inte)
# Returnspin
cdp.fakespin = return_spin(fake_spin_inte)
# Modify data
if spin_inte == ":52@N":
# Set reference data
cdp.fakespin.r2[r20_key] = 6.51945
cdp.fakespin.kex = 13193.82986
cdp.fakespin.kex_err = 2307.09152
phi_ex_rad2_s2 = 93499.92172
phi_ex_err_rad2_s2 = 33233.23039
scaling_rad2_s2 = frequency_to_ppm(frq=1/(2*pi), B0=cdp.spectrometer_frq_list[0], isotope='15N')**2
print scaling_rad2_s2
cdp.fakespin.phi_ex = phi_ex_rad2_s2*scaling_rad2_s2
cdp.fakespin.phi_ex_err = phi_ex_err_rad2_s2*scaling_rad2_s2
print cdp.myspin.ri_data['R1'], cdp.myspin.ri_data_err['R1'], cdp.myspin.r2[r20_key], cdp.myspin.kex, cdp.myspin.phi_ex
print cdp.fakespin.ri_data['R1'], cdp.fakespin.ri_data_err['R1'], cdp.fakespin.r2[r20_key], cdp.fakespin.kex, cdp.fakespin.phi_ex
# Calculate the offset data
theta_spin_dic, Domega_spin_dic, w_eff_spin_dic, dic_key_list = calc_rotating_frame_params(spin=cdp.myspin, spin_id=spin_inte, verbosity=0)
# Save the data in cdp to access it after execution of script.
cdp.myspin.theta_spin_dic = theta_spin_dic
cdp.myspin.w_eff_spin_dic = w_eff_spin_dic
cdp.myspin.dic_key_list = dic_key_list
############################
# First creacte back calculated R2eff data for interpolated plots.
############################
# Return the original structure for frq, offset
spin_lock_nu1 = return_spin_lock_nu1(ref_flag=False)
# Back calculate R2eff data for the set parameters.
cdp.fakespin.back_calc = optimisation.back_calc_r2eff(spin=cdp.fakespin, spin_id=fake_spin_inte, spin_lock_nu1=spin_lock_nu1)
# Prepare list to hold new data
spin_lock_nu1_new = []
# Loop over the structures to generate data
for ei in range(len(spin_lock_nu1)):
# Add a new dimension.
spin_lock_nu1_new.append([])
# Then loop over the spectrometer frequencies.
for mi in range(len(spin_lock_nu1[ei])):
# Add a new dimension.
spin_lock_nu1_new[ei].append([])
# Finally the offsets.
for oi in range(len(spin_lock_nu1[ei][mi])):
# Add a new dimension.
spin_lock_nu1_new[ei][mi].append([])
# No data.
if not len(spin_lock_nu1[ei][mi][oi]):
continue
# Interpolate (adding the extended amount to the end).
for di in range(num_points):
point = (di + 1) * (max(spin_lock_nu1[ei][mi][oi])+extend) / num_points
spin_lock_nu1_new[ei][mi][oi].append(point)
# Intersert field 0
#spin_lock_nu1_new[ei][mi][oi][0] = 0.0
# Convert to a numpy array.
spin_lock_nu1_new[ei][mi][oi] = array(spin_lock_nu1_new[ei][mi][oi], float64)
# Then back calculate R2eff data for the interpolated points.
cdp.myspin.back_calc = optimisation.back_calc_r2eff(spin=cdp.myspin, spin_id=spin_inte, spin_lock_nu1=spin_lock_nu1_new)
# Calculate the offset data, interpolated
theta_spin_dic_inter, Domega_spin_dic_inter, w_eff_spin_dic_inter, dic_key_list_inter = calc_rotating_frame_params(spin=cdp.myspin, spin_id=spin_inte, fields = spin_lock_nu1_new, verbosity=0)
###### Store the data before plotting
# Create a dictionary to hold data
cdp.mydic = collections.OrderedDict()
# Loop over the data structures and save to dictionary
for exp_type, frq, offset, ei, mi, oi in loop_exp_frq_offset(return_indices=True):
# This is not used, but could be used to get Rex.
R1_rho_prime = cdp.myspin.r2[r20_key]
#print R1_rho_prime
# Get R1
R1 = cdp.myspin.ri_data['R1']
R1_err = cdp.myspin.ri_data_err['R1']
# Add to dic
if exp_type not in cdp.mydic:
cdp.mydic[exp_type] = collections.OrderedDict()
if frq not in cdp.mydic[exp_type]:
cdp.mydic[exp_type][frq] = collections.OrderedDict()
if offset not in cdp.mydic[exp_type][frq]:
cdp.mydic[exp_type][frq][offset] = collections.OrderedDict()
# X val
cdp.mydic[exp_type][frq][offset]['point'] = []
cdp.mydic[exp_type][frq][offset]['point_inter'] = []
cdp.mydic[exp_type][frq][offset]['theta'] = []
cdp.mydic[exp_type][frq][offset]['theta_inter'] = []
cdp.mydic[exp_type][frq][offset]['w_eff'] = []
cdp.mydic[exp_type][frq][offset]['w_eff_inter'] = []
# Y val
cdp.mydic[exp_type][frq][offset]['R1_rho'] = []
cdp.mydic[exp_type][frq][offset]['R1_rho_err'] = []
cdp.mydic[exp_type][frq][offset]['R1_rho_bc'] = []
cdp.mydic[exp_type][frq][offset]['R1_rho_inter'] = []
# Y val fake
cdp.mydic[exp_type][frq][offset]['fake_R1_rho'] = []
# Y2 val
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff'] = []
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_err'] = []
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_bc'] = []
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_inter'] = []
# Loop over the original dispersion points.
for point, di in loop_point(exp_type=exp_type, frq=frq, offset=offset, return_indices=True):
param_key = return_param_key_from_data(exp_type=exp_type, frq=frq, offset=offset, point=point)
# X val
cdp.mydic[exp_type][frq][offset]['point'].append(point)
theta = theta_spin_dic[param_key]
cdp.mydic[exp_type][frq][offset]['theta'].append(theta)
w_eff = w_eff_spin_dic[param_key]
cdp.mydic[exp_type][frq][offset]['w_eff'].append(w_eff)
# Average resonance spin_lock_offset
#print Domega_spin_dic[param_key]
# Y val
R1_rho = cdp.myspin.r2eff[param_key]
cdp.mydic[exp_type][frq][offset]['R1_rho'].append(R1_rho)
R1_rho_err = cdp.myspin.r2eff_err[param_key]
cdp.mydic[exp_type][frq][offset]['R1_rho_err'].append(R1_rho_err)
R1_rho_bc = cdp.myspin.r2eff_bc[param_key]
cdp.mydic[exp_type][frq][offset]['R1_rho_bc'].append(R1_rho_bc)
# Y val, fake
fake_R1_rho = cdp.fakespin.back_calc[ei][0][mi][oi][di]
cdp.mydic[exp_type][frq][offset]['fake_R1_rho'].append(fake_R1_rho)
# Y2 val
# Calc R1_rho_R2eff
R1_rho_R2eff = (R1_rho - R1*cos(theta)*cos(theta)) / (sin(theta) * sin(theta))
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff'].append(R1_rho_R2eff)
R1_rho_R2eff_err = (R1_rho_err - R1_err*cos(theta)*cos(theta)) / (sin(theta) * sin(theta))
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_err'].append(R1_rho_R2eff_err)
R1_rho_R2eff_bc = (R1_rho_bc - R1*cos(theta)*cos(theta)) / (sin(theta) * sin(theta))
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_bc'].append(R1_rho_R2eff_bc)
## Loop over the new dispersion points.
for di in range(len(cdp.myspin.back_calc[ei][0][mi][oi])):
point = spin_lock_nu1_new[ei][mi][oi][di]
param_key = return_param_key_from_data(exp_type=exp_type, frq=frq, offset=offset, point=point)
# X val
cdp.mydic[exp_type][frq][offset]['point_inter'].append(point)
theta = theta_spin_dic_inter[param_key]
cdp.mydic[exp_type][frq][offset]['theta_inter'].append(theta)
w_eff = w_eff_spin_dic_inter[param_key]
cdp.mydic[exp_type][frq][offset]['w_eff_inter'].append(w_eff)
# Y val
R1_rho = cdp.myspin.back_calc[ei][0][mi][oi][di]
cdp.mydic[exp_type][frq][offset]['R1_rho_inter'].append(R1_rho)
# Y2 val
# Calc R1_rho_R2eff
R1_rho_R2eff = (R1_rho - R1*cos(theta)*cos(theta)) / (sin(theta) * sin(theta))
cdp.mydic[exp_type][frq][offset]['R1_rho_R2eff_inter'].append(R1_rho_R2eff)
#if oi == 0:
#print exp_type, frq, offset, point, theta, w_eff
####### PLOT ####
## Define labels for plotting
filesave_R1_rho_R2eff = 'R1_rho_R2eff'
filesave_R1_rho = 'R1_rho'
# For writing math in matplotlib, see
# http://matplotlib.org/1.3.1/users/mathtext.html
ylabel_R1_rho = r'R$_{1\rho}$ [rad s$^{-1}$]'
ylabel_R1_rho_R2eff = r'R$_{1\rho, R_{2,eff}}$ [rad s$^{-1}$]'
xlabel_theta = 'Rotating frame tilt angle [rad]'
xlabel_w_eff = r'Effective field in rotating frame [rad s$^{-1}$]'
xlabel_lock = 'Spin-lock field strength [Hz]'
# Set image inches size
img_inch_x = 12
img_inch_y = img_inch_x / 1.6
legend_size = 6
# Plot values in dic
for exptype, frq_dic in cdp.mydic.items():
for frq, offset_dic in frq_dic.items():
for offset, val_dics in offset_dic.items():
# General plot label
graphlabel = "%3.1f_%3.3f_meas"%(frq/1E6, offset)
graphlabel_bc = "%3.1f_%3.3f_bc"%(frq/1E6, offset)
graphlabel_inter = "%3.1f_%3.3f_inter"%(frq/1E6, offset)
graphlabel_fake = "%3.1f_%3.3f_fake"%(frq/1E6, offset)
# Plot 1: R1_rho as function of theta.
plt.figure(1)
line, = plt.plot(val_dics['theta_inter'], val_dics['R1_rho_inter'], '-', label=graphlabel_inter)
plt.errorbar(val_dics['theta'], val_dics['R1_rho'], yerr=val_dics['R1_rho_err'], fmt='o', label=graphlabel, color=line.get_color())
plt.plot(val_dics['theta'], val_dics['R1_rho_bc'], 'D', label=graphlabel_bc, color=line.get_color())
# Plot 2: R1_rho_R2eff as function of w_eff
plt.figure(2)
w_eff2_inter = [x*x for x in val_dics['w_eff_inter']]
w_eff2 = [x*x for x in val_dics['w_eff']]
#line, = plt.plot(w_eff2_inter, val_dics['R1_rho_R2eff_inter'], '-', label=graphlabel_inter)
#plt.errorbar(w_eff2, val_dics['R1_rho_R2eff'], yerr=val_dics['R1_rho_R2eff_err'], fmt='o', label=graphlabel, color=line.get_color())
#plt.plot(w_eff2, val_dics['R1_rho_R2eff_bc'], 'D', label=graphlabel_bc, color=line.get_color())
line, = plt.plot(val_dics['w_eff_inter'], val_dics['R1_rho_R2eff_inter'], '-', label=graphlabel_inter)
plt.errorbar(val_dics['w_eff'], val_dics['R1_rho_R2eff'], yerr=val_dics['R1_rho_R2eff_err'], fmt='o', label=graphlabel, color=line.get_color())
plt.plot(val_dics['w_eff'], val_dics['R1_rho_R2eff_bc'], 'D', label=graphlabel_bc, color=line.get_color())
# Plot 3: R1_rho as function of as function of disp_point, the Spin-lock field strength
plt.figure(3)
line, = plt.plot(val_dics['point_inter'], val_dics['R1_rho_inter'], '-', label=graphlabel_inter)
plt.errorbar(val_dics['point'], val_dics['R1_rho'], yerr=val_dics['R1_rho_err'], fmt='o', label=graphlabel, color=line.get_color())
plt.plot(val_dics['point'], val_dics['R1_rho_bc'], 'D', label=graphlabel_bc, color=line.get_color())
plt.plot(val_dics['point'], val_dics['fake_R1_rho'], '*', label=graphlabel_fake, color=line.get_color())
# Define settings for each graph
# Plot 1: R1_rho as function of theta.
fig1 = plt.figure(1)
plt.xlabel(xlabel_theta)
plt.ylabel(ylabel_R1_rho)
plt.legend(loc='best', prop={'size':legend_size})
plt.grid(True)
#plt.ylim([0,16])
plt.title("%s \n %s as function of %s"%(spin_inte, ylabel_R1_rho, xlabel_theta))
fig1.set_size_inches(img_inch_x, img_inch_y)
plt.savefig("matplotlib_%s_%s_theta_sep.png"%(spin_inte_rep, filesave_R1_rho) )
## Plot 2: R1_rho_R2eff as function of w_eff
fig2 = plt.figure(2)
plt.xlabel(xlabel_w_eff)
plt.ylabel(ylabel_R1_rho_R2eff)
plt.legend(loc='best', prop={'size':legend_size})
plt.grid(True)
#plt.ylim([0,16])
#plt.xlim([0,20000*20000])
plt.xlim([0,20000])
plt.title("%s \n %s as function of %s"%(spin_inte, ylabel_R1_rho_R2eff, xlabel_w_eff))
fig2.set_size_inches(img_inch_x, img_inch_y)
plt.savefig("matplotlib_%s_%s_w_eff.png"%(spin_inte_rep, filesave_R1_rho_R2eff) )
## Plot 3: R1_rho as function of as function of disp_point, the Spin-lock field strength
fig3 = plt.figure(3)
plt.xlabel(xlabel_lock)
plt.ylabel(ylabel_R1_rho)
plt.legend(loc='best', prop={'size':legend_size})
plt.grid(True)
#plt.ylim([0,16])
plt.title("%s \n %s as function of %s"%(spin_inte, ylabel_R1_rho, xlabel_lock))
fig3.set_size_inches(img_inch_x, img_inch_y)
plt.savefig("matplotlib_%s_%s_disp.png"%(spin_inte_rep, filesave_R1_rho_R2eff) )
plt.show()
Bugs ?
Do you get an error with matplotlib about dateutil? Then see Matplotlib_dateutil_bug