Difference between revisions of "Relax disp.spin lock offset+field figure"
Jump to navigation
Jump to search
(→See also: Fix for the category.) |
|||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | {{lowercase title}} | ||
| + | |||
| + | __TOC__ | ||
| + | |||
== Reference to original figure == | == Reference to original figure == | ||
| − | Comparing to Figure 1 and 10 the reference | + | Comparing to Figure 1 and 10 the reference: |
| − | + | ||
| + | * {{#lst:Citations|PalmerMassi06}} | ||
| − | [ | + | [[File:Fig1 Palmer Massi 2006.png|thumb|center|upright=4|Try to reproduce Figure 1.]] |
== Script to produce figure == | == Script to produce figure == | ||
| − | + | ||
| + | {{collapsible script | ||
| + | | type = Python script | ||
| + | | title = Reproduce figure 1 of Palmer and Massi, 2006 using matplotlib. | ||
| + | | lang = python | ||
| + | | script = | ||
############# | ############# | ||
# Made by Troels E. Linnet | # Made by Troels E. Linnet | ||
| Line 222: | Line 232: | ||
# Make Sx | # Make Sx | ||
x, y, z = Sx | x, y, z = Sx | ||
| − | Sx = Arrow3D([-1, x],[0, y], [0, z], mutation_scale=20, lw=1, arrowstyle="- | + | Sx = Arrow3D([-1, x],[0, y], [0, z], mutation_scale=20, lw=1, arrowstyle="-{{!}}>", color="k") |
ax.add_artist(Sx) | ax.add_artist(Sx) | ||
# Make Sy | # Make Sy | ||
x, y, z = Sy | x, y, z = Sy | ||
| − | fSy = Arrow3D([0, x],[1, y], [0, z], mutation_scale=20, lw=1, arrowstyle="- | + | fSy = Arrow3D([0, x],[1, y], [0, z], mutation_scale=20, lw=1, arrowstyle="-{{!}}>", color="k") |
ax.add_artist(fSy) | ax.add_artist(fSy) | ||
# Make Sz | # Make Sz | ||
x, y, z = Sz | x, y, z = Sz | ||
| − | fSz = Arrow3D([0, x],[0, y], [-1, z], mutation_scale=20, lw=1, arrowstyle="- | + | fSz = Arrow3D([0, x],[0, y], [-1, z], mutation_scale=20, lw=1, arrowstyle="-{{!}}>", color="k") |
ax.add_artist(fSz) | ax.add_artist(fSz) | ||
# Make Szp | # Make Szp | ||
x, y, z = Szp | x, y, z = Szp | ||
| − | fSzp = Arrow3D([0, x],[0, y], [0, z], mutation_scale=20, lw=3, arrowstyle="- | + | fSzp = Arrow3D([0, x],[0, y], [0, z], mutation_scale=20, lw=3, arrowstyle="-{{!}}>", color="k") |
ax.add_artist(fSzp) | ax.add_artist(fSzp) | ||
# Make Sxp | # Make Sxp | ||
x, y, z = Sxp | x, y, z = Sxp | ||
| − | fSxp = Arrow3D([0, x],[0, y], [0, z], mutation_scale=20, lw=3, arrowstyle="- | + | fSxp = Arrow3D([0, x],[0, y], [0, z], mutation_scale=20, lw=3, arrowstyle="-{{!}}>", color="k") |
ax.add_artist(fSxp) | ax.add_artist(fSxp) | ||
| Line 257: | Line 267: | ||
# Make we1 | # Make we1 | ||
x, y, z = we1 | x, y, z = we1 | ||
| − | fwe1 = Arrow3D([0, x],[0, y], [0, z], mutation_scale=20, lw=1, arrowstyle="- | + | fwe1 = Arrow3D([0, x],[0, y], [0, z], mutation_scale=20, lw=1, arrowstyle="-{{!}}>", color="b") |
ax.add_artist(fwe1) | ax.add_artist(fwe1) | ||
| Line 271: | Line 281: | ||
# Make we | # Make we | ||
x, y, z = we | x, y, z = we | ||
| − | fwe = Arrow3D([0, x],[0, y], [0, z], mutation_scale=20, lw=1, arrowstyle="- | + | fwe = Arrow3D([0, x],[0, y], [0, z], mutation_scale=20, lw=1, arrowstyle="-{{!}}>", color="r") |
ax.add_artist(fwe) | ax.add_artist(fwe) | ||
| Line 289: | Line 299: | ||
fig.canvas.mpl_connect('button_release_event', update_position) | fig.canvas.mpl_connect('button_release_event', update_position) | ||
plt.show() | plt.show() | ||
| − | + | }} | |
| + | |||
| + | == See also == | ||
| + | |||
| + | [[Category:Relaxation dispersion analysis]] | ||
Latest revision as of 17:35, 6 November 2015
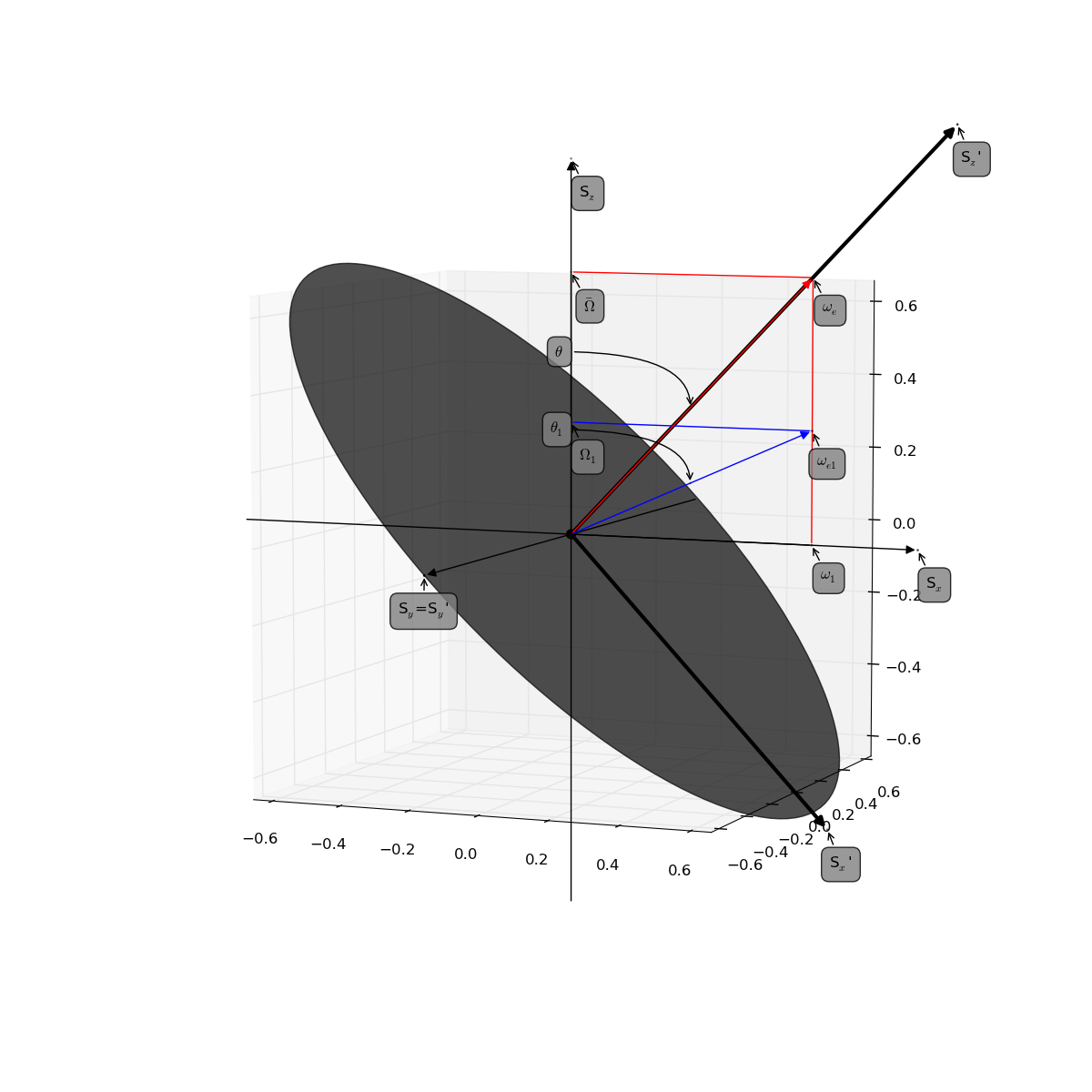
Reference to original figure
Comparing to Figure 1 and 10 the reference:
- Palmer, 3rd, A. G. and Massi, F. (2006). Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem. Rev., 106(5), 1700-1719. (DOI: 10.1021/cr0404287)
Script to produce figure
Python script: Reproduce figure 1 of Palmer and Massi, 2006 using matplotlib.