Difference between revisions of "Tutorial for sorting data stored as numpy to on-resonance R1rho analysis"
Jump to navigation
Jump to search
(Created page with "== Data background == This is data recorded at 600 and 950 MHz.<br> For each spectrometer frequency, the data is saved in np.arrays # one for the residue number, # one for t...") |
|||
| Line 14: | Line 14: | ||
Specifically, the numpy shapes of the data is: | Specifically, the numpy shapes of the data is: | ||
| − | # 600 | + | # For 600 MHz |
## residues (1, 60) | ## residues (1, 60) | ||
## rates (60, 10) | ## rates (60, 10) | ||
| Line 20: | Line 20: | ||
## RFfields (1, 10) | ## RFfields (1, 10) | ||
| − | # 950 | + | # For 950 Mhz |
## residues (1, 61) | ## residues (1, 61) | ||
## rates (61, 19) | ## rates (61, 19) | ||
| Line 26: | Line 26: | ||
## RFfields (1, 19) | ## RFfields (1, 19) | ||
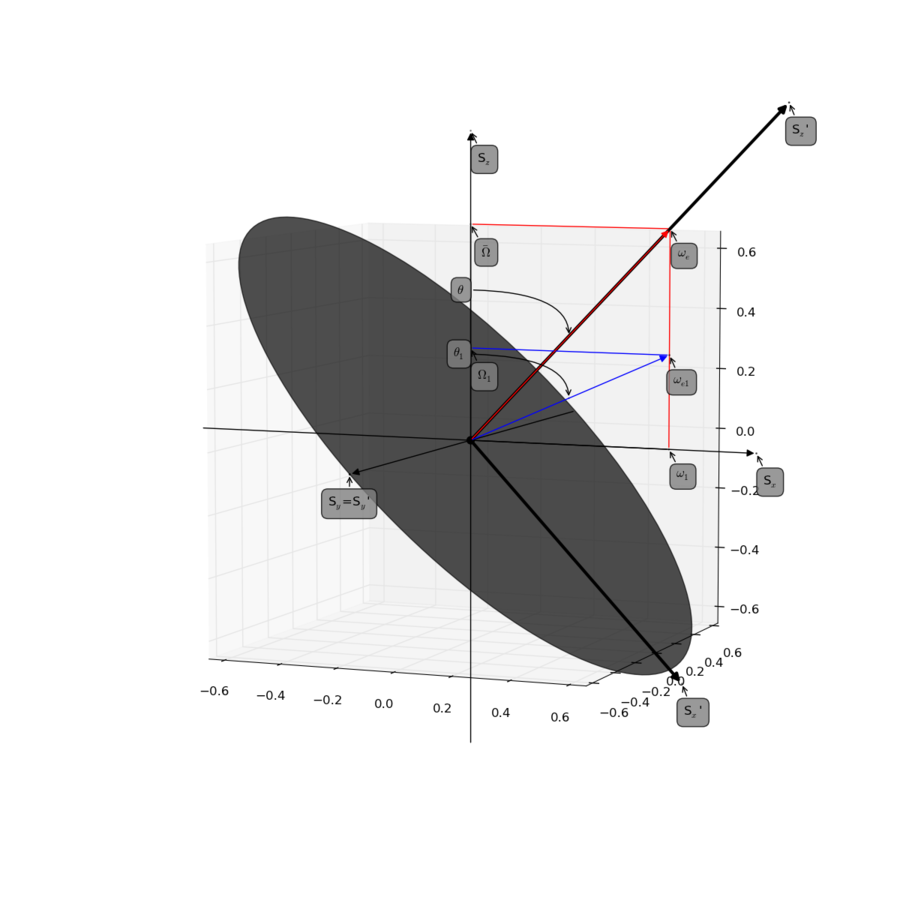
| + | [[File:Fig1 Palmer Massi 2006.png|thumb|center|upright=3|Try to reproduce Figure 1.]] | ||
== See also == | == See also == | ||
[[Category:Tutorials]] | [[Category:Tutorials]] | ||
[[Category:Relaxation dispersion analysis]] | [[Category:Relaxation dispersion analysis]] | ||
Revision as of 22:38, 14 November 2015
Data background
This is data recorded at 600 and 950 MHz.
For each spectrometer frequency, the data is saved in np.arrays
- one for the residue number,
- one for the rates,
- one for the errorbars,
- one for the RF field strength.
They can be retrieved also with scipy's loadmat command.
The experiments are on-resonance R1rho, and the rates are already corrected for the (small) offset effect, using the experimentally determined R1.
Specifically, the numpy shapes of the data is:
- For 600 MHz
- residues (1, 60)
- rates (60, 10)
- errorbars_rate (60, 10)
- RFfields (1, 10)
- For 950 Mhz
- residues (1, 61)
- rates (61, 19)
- errorbars_rate (61, 19)
- RFfields (1, 19)