Difference between revisions of "Tutorial for sorting data stored as numpy to on-resonance R1rho analysis"
| Line 322: | Line 322: | ||
* Inspect which residues should not be included in analysis | * Inspect which residues should not be included in analysis | ||
<source lang="bash"> | <source lang="bash"> | ||
| − | + | sel_ids = [ | |
":4@HN", | ":4@HN", | ||
":5@HN", | ":5@HN", | ||
| Line 331: | Line 331: | ||
] | ] | ||
# De-select for analysis those spins who have bad data | # De-select for analysis those spins who have bad data | ||
| − | for | + | deselet.all() |
| − | print(" | + | for sel_spin in sel_ids: |
| − | + | print("Selecting spin %s"%desel_spin) | |
| + | select.spin(spin_id=desel_spin, change_all=False) | ||
</source> | </source> | ||
Revision as of 23:57, 14 November 2015
Contents
Data background
This is data recorded at 600 and 950 MHz.
This should follow relax-4.0.0.Darwin.dmg installation on a mac, only with GUI.
For each spectrometer frequency, the data is saved in np.arrays
- one for the residue number,
- one for the rates,
- one for the errorbars,
- one for the RF field strength.
They can be retrieved also with scipy's loadmat command.
The experiments are on-resonance R1rho, and the rates are already corrected for the (small) offset effect, using the experimentally determined R1.
Specifically, the numpy shapes of the data is:
- For 600 MHz
- residues (1, 60)
- rates (60, 10)
- errorbars_rate (60, 10)
- RFfields (1, 10)
- For 950 Mhz
- residues (1, 61)
- rates (61, 19)
- errorbars_rate (61, 19)
- RFfields (1, 19)
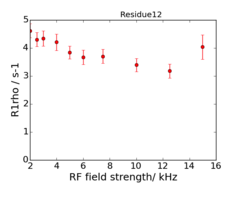
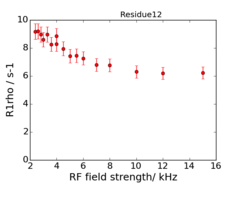
An example of the data at the 2 fields is:
Create data files for relax
First prepare data, by running in python.
python 1_prepare_data.py
1_prepare_data.py
File: 1_prepare_data.py
import os
import scipy as sc
import scipy.io
import numpy as np
# Set path
cwd = os.getcwd()
fields = [600, 950]
file_names = ['residues', 'rates', 'errorbars_rate', 'RFfields']
# Store data in dictionary
all_data = {}
all_data['fields'] = fields
all_data['file_names'] = file_names
# Make list of residues and make unique
all_res = []
# Loop over the experiments, collect all data
for field in fields:
print "\n", field
# Make a dic inside
all_data['%s'%field] = {}
# Construct the path to the data
path = cwd + os.sep + "Archive" + os.sep + "exp_%s"%field + os.sep + "matrices" + os.sep
all_data['%s'%field]['path'] = path
# Collect all filename paths
field_file_name_paths = []
for file_name in file_names:
# Create path name
file_name_path = path + "%s.mat"%file_name
field_file_name_paths.append(file_name_path)
# Load the data
file_name_path_data = sc.io.loadmat(file_name_path)
# Extract as numpy
file_name_path_data_np = file_name_path_data[file_name]
# And store
all_data['%s'%field]['%s'%file_name] = file_name_path_data
all_data['%s'%field]['np_%s'%file_name] = file_name_path_data_np
print file_name, file_name_path_data_np.shape
# Collect residues
if file_name == "residues":
all_res += list(file_name_path_data_np.flatten())
# Store
all_data['%s'%field]['field_file_name_paths'] = field_file_name_paths
# Make list of residues and make unique
all_res_uniq = sorted(list(set(all_res)))
all_data['all_res_uniq'] = all_res_uniq
# Write a sequence file for relax
f = open("residues.txt", "w")
f.write("# Residue_i\n")
for res in all_res_uniq:
f.write("%s\n"%res)
f.close()
f_exp = open("exp_settings.txt", "w")
f_exp.write("# sfrq_MHz RFfield_kHz file_name\n")
# Then write the files for the rates
for field in all_data['fields']:
resis = all_data['%s'%field]['np_residues'][0]
rates = all_data['%s'%field]['np_rates']
errorbars_rate = all_data['%s'%field]['np_errorbars_rate']
RFfields = all_data['%s'%field]['np_RFfields'][0]
print "\nfield: %3.3f"%field
for i, RF_field_strength_kHz in enumerate(RFfields):
#print "RF_field_strength_kHz: %3.3f"%RF_field_strength_kHz
# Generate file name
f_name = "sfrq_%i_MHz_RFfield_%1.3f_kHz.in"%(field, RF_field_strength_kHz)
cur_file = open(f_name, "w")
cur_file.write("# resi rate rate_err\n")
exp_string = "%11.7f %11.7f %s\n"%(field, RF_field_strength_kHz, f_name)
print exp_string,
f_exp.write(exp_string)
for j, resi in enumerate(resis):
rate = rates[j, i]
error = errorbars_rate[j, i]
string = "%4d %11.7f %11.7f\n"%(resi, rate, error)
cur_file.write(string)
cur_file.close()
f_exp.close()
Run analysis in relax GUI
- Start relax
- Then click: "New analysis (Cmd+n)
- Then click icon for "Relaxation dispersion" -> Next
- Just accept name for the pipe
- Open the interpreter: Cmd+p
- Paste in
import os; os.chdir(os.getenv('HOME') + os.sep + 'Desktop' + os.sep + 'temp'); pwd()
- Then do
script(file='2_load_data.py')
- You can scroll through earlier commands with: Cmd+ Arrow up
- When this is done, quit relax
- Then to convert all xmgrace files to png
cd grace
python grace2images.py
This should give the images of the data
- Afterwards you can
- start relax
- Open the interpreter: Cmd+p
- Paste in
import os; os.chdir(os.getenv('HOME') + os.sep + 'Desktop' + os.sep + 'temp'); pwd()
- Close the interpreter
- File -> Open relax state -> "temp_state.bz2"
Here comes an error message we have to investigate. But this can just be closed?
Essentially one need to de-select spin 51 before one select the models for R1rho and then start the analysis.
The script under it can be run by
relax 2_load_data.py -t log.txt
2_load_data.py
File: 2_load_data.py
# relax import
from pipe_control.mol_res_spin import spin_loop
# Test if running as script or through GUI.
is_script = False
if not hasattr(cdp, "pipe_type"):
is_script = True
# We need to create a data pipe, which will tell relax which type of data we are expecting
pipe_name = 'relax_disp'
pipe_bundle = 'relax_disp'
pipe.create(pipe_name, pipe_bundle)
# Minimum: Just read the sequence data, but this misses a lot of information.
sequence.read(file='residues.txt', res_num_col=1)
# Name the spins
for cur_spin, mol_name, resi, resn, spin_id in spin_loop(full_info=True, return_id=True, skip_desel=True):
spin.name(name="HN", spin_id=spin_id)
# Manually force the model to be R2eff, so plotting can be performed later
cur_spin.model = "R2eff"
# Name the isotope for field strength scaling.
spin.isotope(isotope='15N')
# Open the settings file
set_file = open("exp_settings.txt")
set_file_lines = set_file.readlines()
for line in set_file_lines:
if "#" in line[0]:
continue
# Get data
field, RF_field_strength_kHz, f_name = line.split()
# Assign data
spec_id = f_name
relax_disp.exp_type(spectrum_id=spec_id, exp_type='R1rho')
# Set the spectrometer frequency
spectrometer.frequency(id=spec_id, frq=float(field), units='MHz')
# Is in kHz, som convert to Hz
#http://wiki.nmr-relax.com/Relax_disp.spin_lock_offset%2Bfield
#http://www.nmr-relax.com/manual/relax_disp_spin_lock_field.html
disp_frq = float(RF_field_strength_kHz)*1000
# Set The spin-lock field strength, nu1, in Hz
relax_disp.spin_lock_field(spectrum_id=spec_id, field=disp_frq)
# Read the R2eff data
relax_disp.r2eff_read(id=spec_id, file=f_name, dir=None, disp_frq=disp_frq, res_num_col=1, data_col=2, error_col=3)
# Is this necessary? The time, in seconds, of the relaxation period.
#relax_disp.relax_time(spectrum_id=spec_id, time=time_sl)
# Plot data
relax_disp.plot_disp_curves(dir='grace', y_axis='r2_eff', x_axis='disp', num_points=1000, extend_hz=500.0, extend_ppm=500.0, interpolate='disp', force=True)
state.save("temp_state", force=True)
# Do it through script
#if False:
#if True:
if is_script:
# Deselect spin 51, due to weid data point
deselect.spin(spin_id=":51@HN", change_all=False)
import os
from auto_analyses import relax_disp as aa_relax_disp
from lib.dispersion.variables import EXP_TYPE_CPMG_DQ, EXP_TYPE_CPMG_MQ, EXP_TYPE_CPMG_PROTON_MQ, EXP_TYPE_CPMG_PROTON_SQ, EXP_TYPE_CPMG_SQ, EXP_TYPE_CPMG_ZQ, EXP_TYPE_LIST, EXP_TYPE_R1RHO, MODEL_B14_FULL, MODEL_CR72, MODEL_CR72_FULL, MODEL_DPL94, MODEL_IT99, MODEL_LIST_ANALYTIC_CPMG, MODEL_LIST_FULL, MODEL_LIST_NUMERIC_CPMG, MODEL_LM63, MODEL_M61, MODEL_M61B, MODEL_MP05, MODEL_NOREX, MODEL_NS_CPMG_2SITE_3D_FULL, MODEL_NS_CPMG_2SITE_EXPANDED, MODEL_NS_CPMG_2SITE_STAR_FULL, MODEL_NS_R1RHO_2SITE, MODEL_NS_R1RHO_3SITE, MODEL_NS_R1RHO_3SITE_LINEAR, MODEL_PARAMS, MODEL_R2EFF, MODEL_TP02, MODEL_TAP03
# Number of grid search increments. If set to None, then the grid search will be turned off and the default parameter values will be used instead.
#GRID_INC = None
GRID_INC = 21
# The number of Monte Carlo simulations to be used for error analysis at the end of the analysis.
MC_NUM = 500
# Model selection technique.
MODSEL = 'AIC'
result_dir_name = os.getcwd()
# Which models to analyse ?
#MODELS = [MODEL_R2EFF, MODEL_NOREX, MODEL_DPL94, MODEL_TP02, MODEL_TAP03, MODEL_MP05, MODEL_NS_R1RHO_2SITE]
#MODELS = [MODEL_R2EFF, MODEL_NOREX]
# R2EFF shall be skipped here
#MODELS = [MODEL_NOREX]
# http://wiki.nmr-relax.com/M61
MODELS = [MODEL_NOREX, MODEL_M61]
# Fit, instead of read
r1_fit = True
# Go
aa_relax_disp.Relax_disp(pipe_name=pipe_name, pipe_bundle=pipe_bundle, results_dir=result_dir_name, models=MODELS, grid_inc=GRID_INC, mc_sim_num=MC_NUM, modsel=MODSEL, r1_fit=r1_fit)
Run the same analysis in relax through terminal
The analysis can be performed by
python 1_prepare_data.py
relax 2_load_data.py -t log.txt
Following analysis
First make graphs
cd No_Rex/
./grace2images.py -t EPS,PNG
cat r1.out
cat r1rho_prime.out
cd ..
cd M61
./grace2images.py -t PNG
cat kex.out
cat phi_ex.out
cat kex.out | grep "None 12"
cat phi_ex.out | grep "None 12"
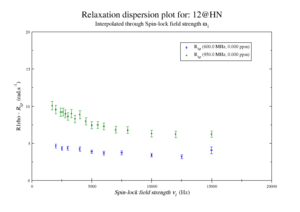
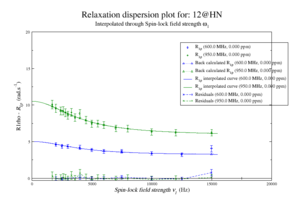
For model M61, spin 12 is:
Resi 12 at 600+950 MHz for M61 model. kex=25000, phi_ex=0.32
Further analysis steps
- Inspect which residues should not be included in analysis
sel_ids = [
":4@HN",
":5@HN",
":6@HN",
":7@HN",
":10@HN",
":11@HN",
]
# De-select for analysis those spins who have bad data
deselet.all()
for sel_spin in sel_ids:
print("Selecting spin %s"%desel_spin)
select.spin(spin_id=desel_spin, change_all=False)
- Inspect which residues should be analysed together for a clustered/global fit.
cluster_ids = [
":13@N",
":15@N",
":16@N",
":25@N"]
# Cluster spins
for curspin in cluster_ids:
print("Adding spin %s to cluster"%curspin)
relax_disp.cluster('model_cluster', curspin)
- Run analysis again