Difference between revisions of "Chi2 surface plot"
(Switched to the {{relax source}} template for repository links. The <include> tags are no longer functional. And the files are now differently named, so most link are dead.) |
|||
| (25 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | __TOC__ | ||
| + | |||
== Plot surface == | == Plot surface == | ||
=== 1_create_surface_data.py === | === 1_create_surface_data.py === | ||
| − | + | {{relax source|path=test_suite/shared_data/dispersion/KTeilum_FMPoulsen_MAkke_2006/surface_chi2_clustered_fitting/1_create_surface_data.py|text=Get file}} | |
<include | <include | ||
highlight="python" | highlight="python" | ||
| Line 9: | Line 11: | ||
=== 1_surface_chi2.txt === | === 1_surface_chi2.txt === | ||
| − | + | {{relax source|path=test_suite/shared_data/dispersion/KTeilum_FMPoulsen_MAkke_2006/surface_chi2_clustered_fitting/1_surface_chi2.txt|text=Get file}} | |
<include | <include | ||
highlight="text" | highlight="text" | ||
| Line 17: | Line 19: | ||
<source lang="text"> | <source lang="text"> | ||
... | ... | ||
| − | </ | + | </source> |
| + | |||
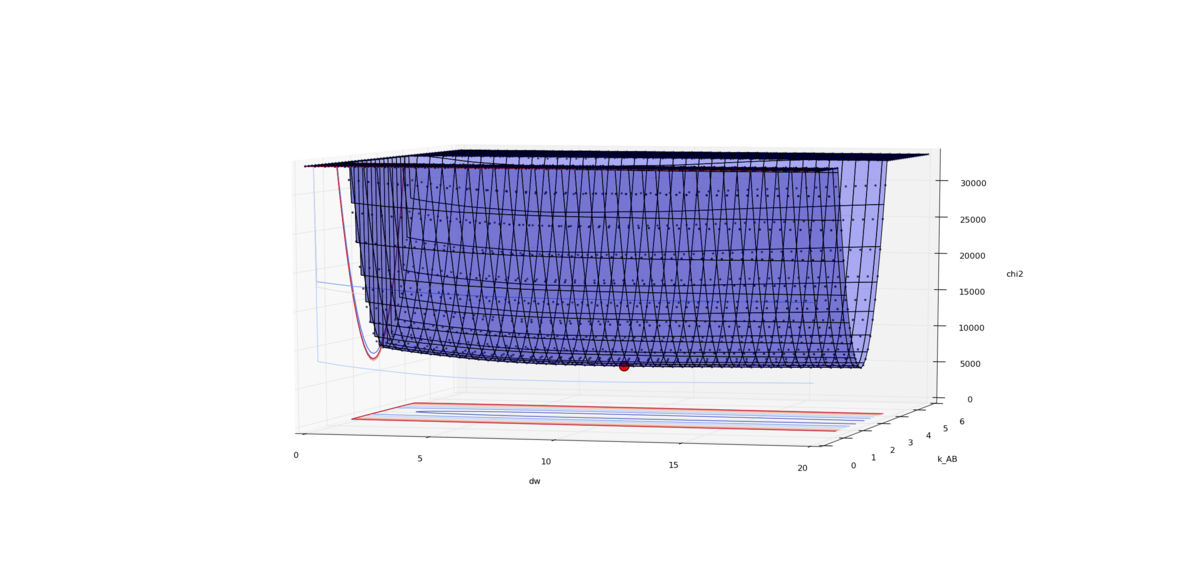
| + | === 2_make_surface_plot.py === | ||
| + | [[File:2 make surface plot.png|thumb|upright=4|Chi2 surface plot]] | ||
| + | |||
| + | {{relax source|path=test_suite/shared_data/dispersion/KTeilum_FMPoulsen_MAkke_2006/surface_chi2_clustered_fitting/2_make_surface_plot.py|text=Get file}} | ||
| + | <include | ||
| + | highlight="python" | ||
| + | src="http://svn.gna.org/viewcvs/*checkout*/relax/trunk/test_suite/shared_data/dispersion/KTeilum_FMPoulsen_MAkke_2006/surface_chi2_clustered_fitting/2_make_surface_plot.py?revision=HEAD" | ||
| + | /> | ||
| + | |||
| + | == Compare dx map == | ||
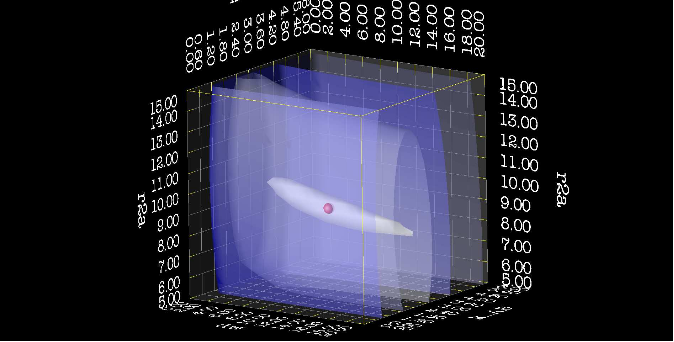
| + | === TSMFK01_dx_map.py === | ||
| + | [[File:Dx map 1.png|thumb|upright=4|dx map of chi2 surface, from corner]] | ||
| + | |||
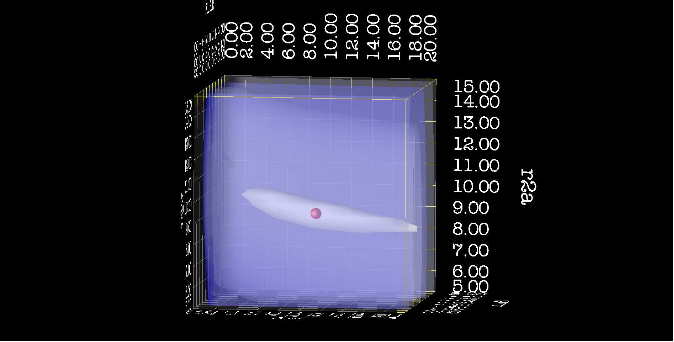
| + | [[File:Map 2.png|thumb|upright=4|dx map of chi2 surface, from side]] | ||
| + | |||
| + | {{relax source|path=test_suite/shared_data/dispersion/KTeilum_FMPoulsen_MAkke_2006/surface_chi2_clustered_fitting/TSMFK01_dx_map.py|text=Get file}} | ||
| + | <include | ||
| + | highlight="python" | ||
| + | src="http://svn.gna.org/viewcvs/*checkout*/relax/trunk/test_suite/shared_data/dispersion/KTeilum_FMPoulsen_MAkke_2006/surface_chi2_clustered_fitting/TSMFK01_dx_map.py?revision=HEAD" | ||
| + | /> | ||
| + | |||
| + | == Create simulated dispersion plots == | ||
| + | |||
| + | {{relax source|path=test_suite/shared_data/dispersion/KTeilum_FMPoulsen_MAkke_2006/surface_chi2_clustered_fitting/3_simulate_graphs.py|text=Get file}} | ||
| + | <include | ||
| + | highlight="python" | ||
| + | src="{{relax url|path=test_suite/shared_data/dispersion/KTeilum_FMPoulsen_MAkke_2006/surface_chi2_clustered_fitting/3_simulate_graphs.py|view=raw}}" | ||
| + | /> | ||
| + | |||
| + | === Gallery === | ||
| + | <gallery> | ||
| + | File:Disp 1002 N.png|dw=10.405 ,k_AB=2.414 ,chi2=6722.833 | ||
| + | File:Disp 1003 N.png|dw=0 ,k_AB=0 ,chi2=98892.958 | ||
| + | File:Disp 1004 N.png|dw=0 ,k_AB=1 ,chi2=72102.777 | ||
| + | File:Disp 1005 N.png|dw=0 ,k_AB=2 ,chi2=182636.657 | ||
| + | File:Disp 1006 N.png|dw=0 ,k_AB=3 ,chi2=430494.6 | ||
| + | File:Disp 1007 N.png|dw=0 ,k_AB=4 ,chi2=815676.604 | ||
| + | File:Disp 1008 N.png|dw=0 ,k_AB=5 ,chi2=1338182.67 | ||
| + | File:Disp 1009 N.png|dw=0 ,k_AB=6 ,chi2=1998012.798 | ||
| + | File:Disp 1010 N.png|dw=3.333 ,k_AB=0 ,chi2=98892.958 | ||
| + | File:Disp 1011 N.png|dw=3.333 ,k_AB=1 ,chi2=38960.501 | ||
| + | File:Disp 1012 N.png|dw=3.333 ,k_AB=2 ,chi2=10243.151 | ||
| + | File:Disp 1013 N.png|dw=3.333 ,k_AB=3 ,chi2=12740.906 | ||
| + | File:Disp 1014 N.png|dw=3.333 ,k_AB=4 ,chi2=46453.768 | ||
| + | File:Disp 1015 N.png|dw=3.333 ,k_AB=5 ,chi2=111381.736 | ||
| + | File:Disp 1016 N.png|dw=3.333 ,k_AB=6 ,chi2=207524.811 | ||
| + | File:Disp 1017 N.png|dw=6.667 ,k_AB=0 ,chi2=98892.958 | ||
| + | File:Disp 1018 N.png|dw=6.667 ,k_AB=1 ,chi2=38516.101 | ||
| + | File:Disp 1019 N.png|dw=6.667 ,k_AB=2 ,chi2=9596.92 | ||
| + | File:Disp 1020 N.png|dw=6.667 ,k_AB=3 ,chi2=12135.415 | ||
| + | File:Disp 1021 N.png|dw=6.667 ,k_AB=4 ,chi2=46131.586 | ||
| + | File:Disp 1022 N.png|dw=6.667 ,k_AB=5 ,chi2=111585.433 | ||
| + | File:Disp 1023 N.png|dw=6.667 ,k_AB=6 ,chi2=208496.955 | ||
| + | File:Disp 1024 N.png|dw=10 ,k_AB=0 ,chi2=98892.958 | ||
| + | File:Disp 1025 N.png|dw=10 ,k_AB=1 ,chi2=38338.21 | ||
| + | File:Disp 1026 N.png|dw=10 ,k_AB=2 ,chi2=9428.183 | ||
| + | File:Disp 1027 N.png|dw=10 ,k_AB=3 ,chi2=12162.877 | ||
| + | File:Disp 1028 N.png|dw=10 ,k_AB=4 ,chi2=46542.291 | ||
| + | File:Disp 1029 N.png|dw=10 ,k_AB=5 ,chi2=112566.427 | ||
| + | File:Disp 1030 N.png|dw=10 ,k_AB=6 ,chi2=210235.283 | ||
| + | File:Disp 1031 N.png|dw=13.333 ,k_AB=0 ,chi2=98892.958 | ||
| + | File:Disp 1032 N.png|dw=13.333 ,k_AB=1 ,chi2=38274.865 | ||
| + | File:Disp 1033 N.png|dw=13.333 ,k_AB=2 ,chi2=9441.93 | ||
| + | File:Disp 1034 N.png|dw=13.333 ,k_AB=3 ,chi2=12394.152 | ||
| + | File:Disp 1035 N.png|dw=13.333 ,k_AB=4 ,chi2=47131.531 | ||
| + | File:Disp 1036 N.png|dw=13.333 ,k_AB=5 ,chi2=113654.068 | ||
| + | File:Disp 1037 N.png|dw=13.333 ,k_AB=6 ,chi2=211961.762 | ||
| + | File:Disp 1038 N.png|dw=16.667 ,k_AB=0 ,chi2=98892.958 | ||
| + | File:Disp 1039 N.png|dw=16.667 ,k_AB=1 ,chi2=38274.349 | ||
| + | File:Disp 1040 N.png|dw=16.667 ,k_AB=2 ,chi2=9547.429 | ||
| + | File:Disp 1041 N.png|dw=16.667 ,k_AB=3 ,chi2=12712.196 | ||
| + | File:Disp 1042 N.png|dw=16.667 ,k_AB=4 ,chi2=47768.651 | ||
| + | File:Disp 1043 N.png|dw=16.667 ,k_AB=5 ,chi2=114716.793 | ||
| + | File:Disp 1044 N.png|dw=16.667 ,k_AB=6 ,chi2=213556.624 | ||
| + | File:Disp 1045 N.png|dw=20 ,k_AB=0 ,chi2=98892.958 | ||
| + | File:Disp 1046 N.png|dw=20 ,k_AB=1 ,chi2=38267.144 | ||
| + | File:Disp 1047 N.png|dw=20 ,k_AB=2 ,chi2=9643.821 | ||
| + | File:Disp 1048 N.png|dw=20 ,k_AB=3 ,chi2=13022.989 | ||
| + | File:Disp 1049 N.png|dw=20 ,k_AB=4 ,chi2=48404.648 | ||
| + | File:Disp 1050 N.png|dw=20 ,k_AB=5 ,chi2=115788.799 | ||
| + | |||
| + | </gallery> | ||
== See also == | == See also == | ||
[[Category:Matplotlib]] | [[Category:Matplotlib]] | ||
[[Category:Relaxation_dispersion_analysis]] | [[Category:Relaxation_dispersion_analysis]] | ||
Latest revision as of 12:33, 27 October 2017
Contents
Plot surface
1_create_surface_data.py
Get file <include highlight="python" src="http://svn.gna.org/viewcvs/*checkout*/relax/trunk/test_suite/shared_data/dispersion/KTeilum_FMPoulsen_MAkke_2006/surface_chi2_clustered_fitting/1_create_surface_data.py?revision=HEAD" />
1_surface_chi2.txt
Get file <include highlight="text" lines="0-10" src="http://svn.gna.org/viewcvs/*checkout*/relax/trunk/test_suite/shared_data/dispersion/KTeilum_FMPoulsen_MAkke_2006/surface_chi2_clustered_fitting/1_surface_chi2.txt?revision=HEAD" />
...
2_make_surface_plot.py
Get file <include highlight="python" src="http://svn.gna.org/viewcvs/*checkout*/relax/trunk/test_suite/shared_data/dispersion/KTeilum_FMPoulsen_MAkke_2006/surface_chi2_clustered_fitting/2_make_surface_plot.py?revision=HEAD" />
Compare dx map
TSMFK01_dx_map.py
Get file <include highlight="python" src="http://svn.gna.org/viewcvs/*checkout*/relax/trunk/test_suite/shared_data/dispersion/KTeilum_FMPoulsen_MAkke_2006/surface_chi2_clustered_fitting/TSMFK01_dx_map.py?revision=HEAD" />
Create simulated dispersion plots
Get file <include highlight="python" src="https://sourceforge.net/p/nmr-relax/code/ci/master/tree/test_suite/shared_data/dispersion/KTeilum_FMPoulsen_MAkke_2006/surface_chi2_clustered_fitting/3_simulate_graphs.py?format=raw" />