Difference between revisions of "Tutorial for sorting data stored as numpy to on-resonance R1rho analysis"
(Created page with "== Data background == This is data recorded at 600 and 950 MHz.<br> For each spectrometer frequency, the data is saved in np.arrays # one for the residue number, # one for t...") |
(Forced creation of a TOC - this will improve the formatting on the main page 'Did you know...' section.) |
||
| (60 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| + | __TOC__ | ||
| + | |||
== Data background == | == Data background == | ||
This is data recorded at 600 and 950 MHz.<br> | This is data recorded at 600 and 950 MHz.<br> | ||
| + | This should follow <code>relax-4.0.1.Darwin.dmg</code> installation on a Mac, only with GUI. | ||
| − | For each spectrometer frequency, the data is saved in np.arrays | + | For each spectrometer frequency, the data is saved in <code>np.arrays</code> |
# one for the residue number, | # one for the residue number, | ||
# one for the rates, | # one for the rates, | ||
| Line 10: | Line 13: | ||
They can be retrieved also with scipy's loadmat command. | They can be retrieved also with scipy's loadmat command. | ||
| − | The experiments are on-resonance R1rho, and the rates are already corrected for the (small) offset effect, using the experimentally determined R1. | + | The experiments are on-resonance {{:R1rho}}, and the rates are already corrected for the (small) offset effect, using the experimentally determined {{:R1}}. |
Specifically, the numpy shapes of the data is: | Specifically, the numpy shapes of the data is: | ||
| − | # 600 | + | # For 600 MHz |
## residues (1, 60) | ## residues (1, 60) | ||
## rates (60, 10) | ## rates (60, 10) | ||
| Line 20: | Line 23: | ||
## RFfields (1, 10) | ## RFfields (1, 10) | ||
| − | # 950 | + | # For 950 Mhz |
## residues (1, 61) | ## residues (1, 61) | ||
## rates (61, 19) | ## rates (61, 19) | ||
| Line 26: | Line 29: | ||
## RFfields (1, 19) | ## RFfields (1, 19) | ||
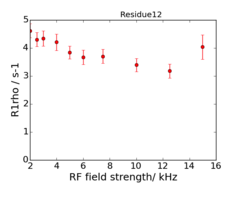
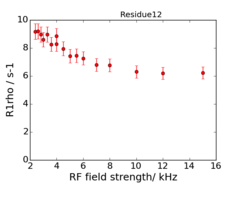
| + | '''For the 950 MHz, the 4000 Hz dispersion point is replicated.''' | ||
| + | |||
| + | |||
| + | An example of the data at the 2 fields is: | ||
| + | <gallery widths=240px heights=240px > | ||
| + | File:Residue12 600.png|600 MHz | ||
| + | File:Residue12 950.png|950 MHz | ||
| + | </gallery> | ||
| + | |||
| + | == Create data files for relax == | ||
| + | First prepare data, by running in Python: | ||
| + | <source lang="bash"> | ||
| + | python 1_prepare_data.py | ||
| + | </source> | ||
| + | |||
| + | The script is: | ||
| + | |||
| + | {{collapsible script | ||
| + | | type = Python 2/3 script | ||
| + | | title = The <code>1_prepare_data.py</code> script. | ||
| + | | lang = python | ||
| + | | script = | ||
| + | import os | ||
| + | from scipy.io import loadmat | ||
| + | import numpy as np | ||
| + | |||
| + | # Set path | ||
| + | #cwd = status.install_path+os.sep+'test_suite'+os.sep+'shared_data'+os.sep+'dispersion'+os.sep+'Paul_Schanda_2015_Nov' | ||
| + | cwd = os.getcwd() | ||
| + | outdir = cwd + os.sep | ||
| + | |||
| + | fields = [600, 950] | ||
| + | file_names = ['residues', 'rates', 'errorbars_rate', 'RFfields'] | ||
| + | |||
| + | # Store data in dictionary | ||
| + | all_data = {} | ||
| + | all_data['fields'] = fields | ||
| + | all_data['file_names'] = file_names | ||
| + | |||
| + | # Make list of residues and make unique | ||
| + | all_res = [] | ||
| + | |||
| + | # Loop over the experiments, collect all data | ||
| + | for field in fields: | ||
| + | print("%s"%field,) | ||
| + | |||
| + | # Make a dic inside | ||
| + | all_data['%s'%field] = {} | ||
| + | |||
| + | # Construct the path to the data | ||
| + | path = cwd + os.sep | ||
| + | |||
| + | # Collect all filename paths | ||
| + | field_file_name_paths = [] | ||
| + | for file_name in file_names: | ||
| + | # Create path name | ||
| + | file_name_path = path + "%s_%s.mat"%(field, file_name) | ||
| + | field_file_name_paths.append(file_name_path) | ||
| + | |||
| + | # Load the data | ||
| + | file_name_path_data = loadmat(file_name_path) | ||
| + | # Extract as numpy | ||
| + | file_name_path_data_np = file_name_path_data[file_name] | ||
| + | # And store | ||
| + | all_data['%s'%field]['%s'%file_name] = file_name_path_data | ||
| + | all_data['%s'%field]['np_%s'%file_name] = file_name_path_data_np | ||
| + | |||
| + | print(file_name, file_name_path_data_np.shape) | ||
| + | |||
| + | # Collect residues | ||
| + | if file_name == "residues": | ||
| + | all_res += list(file_name_path_data_np.flatten()) | ||
| + | |||
| + | # Store | ||
| + | all_data['%s'%field]['field_file_name_paths'] = field_file_name_paths | ||
| + | |||
| + | |||
| + | # Make list of residues and make unique | ||
| + | all_res_uniq = sorted(list(set(all_res))) | ||
| + | all_data['all_res_uniq'] = all_res_uniq | ||
| + | |||
| + | # Write a sequence file for relax | ||
| + | f = open(outdir + "residues.txt", "w") | ||
| + | f.write("# Residue_i\n") | ||
| + | for res in all_res_uniq: | ||
| + | f.write("%s\n"%res) | ||
| + | f.close() | ||
| + | |||
| + | f_exp = open(outdir + "exp_settings.txt", "w") | ||
| + | f_exp.write("# sfrq_MHz RFfield_kHz file_name\n") | ||
| + | |||
| + | # Then write the files for the rates | ||
| + | k = 1 | ||
| + | for field in all_data['fields']: | ||
| + | resis = all_data['%s'%field]['np_residues'][0] | ||
| + | rates = all_data['%s'%field]['np_rates'] | ||
| + | errorbars_rate = all_data['%s'%field]['np_errorbars_rate'] | ||
| + | RFfields = all_data['%s'%field]['np_RFfields'][0] | ||
| + | |||
| + | print("\nfield: %3.3f"%field) | ||
| + | for i, RF_field_strength_kHz in enumerate(RFfields): | ||
| + | #print "RF_field_strength_kHz: %3.3f"%RF_field_strength_kHz | ||
| + | # Generate file name | ||
| + | f_name = outdir + "sfrq_%i_MHz_RFfield_%1.3f_kHz_%03d.in"%(field, RF_field_strength_kHz, k) | ||
| + | cur_file = open(f_name, "w") | ||
| + | cur_file.write("# resi rate rate_err\n") | ||
| + | |||
| + | exp_string = "%11.7f %11.7f %s\n"%(field, RF_field_strength_kHz, f_name) | ||
| + | print("%s"%exp_string,) | ||
| + | f_exp.write(exp_string) | ||
| + | |||
| + | for j, resi in enumerate(resis): | ||
| + | rate = rates[j, i] | ||
| + | error = errorbars_rate[j, i] | ||
| + | string = "%4d %11.7f %11.7f\n"%(resi, rate, error) | ||
| + | cur_file.write(string) | ||
| + | |||
| + | cur_file.close() | ||
| + | k += 1 | ||
| + | |||
| + | f_exp.close() | ||
| + | }} | ||
| + | |||
| + | == Run analysis in relax GUI == | ||
| + | * Start relax | ||
| + | * Then click: {{button|new analysis}} or {{key press|cmd|n}} | ||
| + | * Then click icon for {{button|Relaxation dispersion}} {{next}} {{button|Next}} | ||
| + | * Just accept name for the pipe | ||
| + | |||
| + | * Open the interpreter: {{key press|cmd|p}} | ||
| + | * Paste in | ||
| + | <source lang="python"> | ||
| + | import os; os.chdir(os.getenv('HOME') + os.sep + 'Desktop' + os.sep + 'temp'); pwd() | ||
| + | </source> | ||
| + | * Then do | ||
| + | <source lang="python"> | ||
| + | script(file='2_load_data.py') | ||
| + | </source> | ||
| + | * ( You can scroll through earlier commands with: {{key press|cmd|up}} ) | ||
| + | * Close the Grace <code>Results viewer</code> window | ||
| + | * Close the interpreter window | ||
| + | |||
| + | Now save the current state! This saved state can now be loaded just before an analysis, and contain all setup and data. | ||
| + | * {{menu|File|Save as (Shift+Ctrl+S)}} {{next}} Save as <code>before_analysis.bz2</code> | ||
| + | |||
| + | Now '''Quit relax''', and start relax again.<br> | ||
| + | Go to: {{menu|File|Open relax state (Ctrl+O)}} {{next}} Locate <code>before_analysis.bz2</code> | ||
| + | |||
| + | In the GUI, select: | ||
| + | * For models, only select: <code>No Rex</code> and <code>M61</code> | ||
| + | * {{:R1}} parameter optimisation to False, as this '''on-resonance''' data does not use {{:R1}}. | ||
| + | * The number of Monte-Carlo simulations to <code>3</code>. (The minimum number before relax refuse to run). | ||
| + | * Let other things be standard, and click execute | ||
| + | |||
| + | {{note|The number of Monte-Carlo simulations is set to '''3'''. This means that relax will do the analysis, and the fitted parameters will be correct, but the standard error of the parameters will be wrong.}} | ||
| + | |||
| + | The reason for this is, that the data "naturally"? for some spins contains measurements which are "doubtful". <br> | ||
| + | When relax is performing Monte-Carlo simulations, all the data is first copied x times to the number of Monte-Carlo simulations, and each {{:R2eff}} point on the graph | ||
| + | is modified randomly by a gaussian noise with a width described by the associated errors. | ||
| + | |||
| + | For spins which contains measurements which are "doubtful", relax will be spending "very long time" trying to fit a meaningful model. <br> | ||
| + | This "time" is poorly spend on "bad data". | ||
| + | |||
| + | Rather, one should first try to | ||
| + | * analyse the data quickly | ||
| + | * make the graphs | ||
| + | * examine which spins should be deselected | ||
| + | * and then rerun the analysis with a higher number of monte-carlo simulations | ||
| + | |||
| + | After this comes the part with global fit/clustered analysis. | ||
| + | * analyse the data again | ||
| + | * Select which spins should be included in a global fit/clustered analysis | ||
| + | * and then rerun with analysis | ||
| + | |||
| + | === The script=== | ||
| + | |||
| + | {{collapsible script | ||
| + | | type = relax script | ||
| + | | title = The <code>2_load_data.py</code> script. | ||
| + | | lang = python | ||
| + | | script = | ||
| + | ############################################################################### | ||
| + | # # | ||
| + | # Copyright (C) 2015 Troels E. Linnet # | ||
| + | # # | ||
| + | # This file is part of the program relax (http://www.nmr-relax.com). # | ||
| + | # # | ||
| + | # This program is free software: you can redistribute it and/or modify # | ||
| + | # it under the terms of the GNU General Public License as published by # | ||
| + | # the Free Software Foundation, either version 3 of the License, or # | ||
| + | # (at your option) any later version. # | ||
| + | # # | ||
| + | # This program is distributed in the hope that it will be useful, # | ||
| + | # but WITHOUT ANY WARRANTY; without even the implied warranty of # | ||
| + | # MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the # | ||
| + | # GNU General Public License for more details. # | ||
| + | # # | ||
| + | # You should have received a copy of the GNU General Public License # | ||
| + | # along with this program. If not, see <http://www.gnu.org/licenses/>. # | ||
| + | # # | ||
| + | ############################################################################### | ||
| + | # Test if running as script or through GUI. | ||
| + | is_script = False | ||
| + | if not hasattr(cdp, "pipe_type"): | ||
| + | is_script = True | ||
| + | # We need to create a data pipe, which will tell relax which type of data we are expecting | ||
| + | pipe_name = 'relax_disp' | ||
| + | pipe_bundle = 'relax_disp' | ||
| + | pipe.create(pipe_name, pipe_bundle) | ||
| + | |||
| + | # Minimum: Just read the sequence data, but this misses a lot of information. | ||
| + | sequence.read(file='residues.txt', res_num_col=1) | ||
| + | |||
| + | # Open the settings file | ||
| + | set_file = open("exp_settings.txt") | ||
| + | set_file_lines = set_file.readlines() | ||
| + | |||
| + | for line in set_file_lines: | ||
| + | if "#" in line[0]: | ||
| + | continue | ||
| + | |||
| + | # Get data | ||
| + | field, RF_field_strength_kHz, f_name = line.split() | ||
| + | |||
| + | # Assign data | ||
| + | spec_id = f_name | ||
| + | relax_disp.exp_type(spectrum_id=spec_id, exp_type='R1rho') | ||
| + | |||
| + | # Set the spectrometer frequency | ||
| + | spectrometer.frequency(id=spec_id, frq=float(field), units='MHz') | ||
| + | |||
| + | # Is in kHz, som convert to Hz | ||
| + | #http://wiki.nmr-relax.com/Relax_disp.spin_lock_offset%2Bfield | ||
| + | #http://www.nmr-relax.com/manual/relax_disp_spin_lock_field.html | ||
| + | disp_frq = float(RF_field_strength_kHz)*1000 | ||
| + | |||
| + | # Set The spin-lock field strength, nu1, in Hz | ||
| + | relax_disp.spin_lock_field(spectrum_id=spec_id, field=disp_frq) | ||
| + | |||
| + | # Read the R2eff data | ||
| + | relax_disp.r2eff_read(id=spec_id, file=f_name, dir=None, disp_frq=disp_frq, res_num_col=1, data_col=2, error_col=3) | ||
| + | |||
| + | # Is this necessary? The time, in seconds, of the relaxation period. | ||
| + | #relax_disp.relax_time(spectrum_id=spec_id, time=time_sl) | ||
| + | |||
| + | |||
| + | # Name the isotope for field strength scaling. | ||
| + | spin.isotope(isotope='15N') | ||
| + | relax_disp.select_model(model='R2eff') | ||
| + | |||
| + | # Plot data | ||
| + | relax_disp.plot_disp_curves(dir='grace', y_axis='r2_eff', x_axis='disp', num_points=1000, extend_hz=500.0, extend_ppm=500.0, interpolate='disp', force=True) | ||
| + | |||
| + | state.save("temp_state", force=True) | ||
| + | |||
| + | # Do it through script | ||
| + | if False: | ||
| + | #if True: | ||
| + | #if is_script: | ||
| + | import os | ||
| + | from auto_analyses import relax_disp as aa_relax_disp | ||
| + | from lib.dispersion.variables import EXP_TYPE_CPMG_DQ, EXP_TYPE_CPMG_MQ, EXP_TYPE_CPMG_PROTON_MQ, EXP_TYPE_CPMG_PROTON_SQ, EXP_TYPE_CPMG_SQ, EXP_TYPE_CPMG_ZQ, EXP_TYPE_LIST, EXP_TYPE_R1RHO, MODEL_B14_FULL, MODEL_CR72, MODEL_CR72_FULL, MODEL_DPL94, MODEL_IT99, MODEL_LIST_ANALYTIC_CPMG, MODEL_LIST_FULL, MODEL_LIST_NUMERIC_CPMG, MODEL_LM63, MODEL_M61, MODEL_M61B, MODEL_MP05, MODEL_NOREX, MODEL_NS_CPMG_2SITE_3D_FULL, MODEL_NS_CPMG_2SITE_EXPANDED, MODEL_NS_CPMG_2SITE_STAR_FULL, MODEL_NS_R1RHO_2SITE, MODEL_NS_R1RHO_3SITE, MODEL_NS_R1RHO_3SITE_LINEAR, MODEL_PARAMS, MODEL_R2EFF, MODEL_TP02, MODEL_TAP03 | ||
| + | # Number of grid search increments. If set to None, then the grid search will be turned off and the default parameter values will be used instead. | ||
| + | #GRID_INC = None | ||
| + | GRID_INC = 21 | ||
| + | # The number of Monte Carlo simulations to be used for error analysis at the end of the analysis. | ||
| + | MC_NUM = 3 | ||
| + | # Model selection technique. | ||
| + | MODSEL = 'AIC' | ||
| + | result_dir_name = os.getcwd() + os.sep + "results" | ||
| + | # Which models to analyse ? | ||
| + | #MODELS = [MODEL_R2EFF, MODEL_NOREX, MODEL_DPL94, MODEL_TP02, MODEL_TAP03, MODEL_MP05, MODEL_NS_R1RHO_2SITE] | ||
| + | #MODELS = [MODEL_R2EFF, MODEL_NOREX] | ||
| + | # R2EFF shall be skipped here | ||
| + | #MODELS = [MODEL_NOREX] | ||
| + | # http://wiki.nmr-relax.com/M61 | ||
| + | MODELS = [MODEL_NOREX, MODEL_M61] | ||
| + | |||
| + | # Fit, instead of read | ||
| + | #r1_fit = True | ||
| + | r1_fit = False | ||
| + | |||
| + | # Set the initial guess from the minimum R2eff point | ||
| + | set_grid_r20=True | ||
| + | |||
| + | """ | ||
| + | @keyword pipe_name: The name of the data pipe containing all of the data for the analysis. | ||
| + | @type pipe_name: str | ||
| + | @keyword pipe_bundle: The data pipe bundle to associate all spawned data pipes with. | ||
| + | @type pipe_bundle: str | ||
| + | @keyword results_dir: The directory where results files are saved. | ||
| + | @type results_dir: str | ||
| + | @keyword models: The list of relaxation dispersion models to optimise. | ||
| + | @type models: list of str | ||
| + | @keyword grid_inc: Number of grid search increments. If set to None, then the grid search will be turned off and the default parameter values will be used instead. | ||
| + | @type grid_inc: int or None | ||
| + | @keyword mc_sim_num: The number of Monte Carlo simulations to be used for error analysis at the end of the analysis. | ||
| + | @type mc_sim_num: int | ||
| + | @keyword exp_mc_sim_num: The number of Monte Carlo simulations for the error analysis in the 'R2eff' model when exponential curves are fitted. This defaults to the value of the mc_sim_num argument when not given. When set to '-1', the R2eff errors are estimated from the Covariance matrix. For the 2-point fixed-time calculation for the 'R2eff' model, this argument is ignored. | ||
| + | @type exp_mc_sim_num: int or None | ||
| + | @keyword modsel: The model selection technique to use in the analysis to determine which model is the best for each spin cluster. This can currently be one of 'AIC', 'AICc', and 'BIC'. | ||
| + | @type modsel: str | ||
| + | @keyword pre_run_dir: The optional directory containing the dispersion auto-analysis results from a previous run. The optimised parameters from these previous results will be used as the starting point for optimisation rather than performing a grid search. This is essential for when large spin clusters are specified, as a grid search becomes prohibitively expensive with clusters of three or more spins. At some point a RelaxError will occur because the grid search is impossibly large. For the cluster specific parameters, i.e. the populations of the states and the exchange parameters, an average value will be used as the starting point. For all other parameters, the R20 values for each spin and magnetic field, as well as the parameters related to the chemical shift difference dw, the optimised values of the previous run will be directly copied. | ||
| + | @type pre_run_dir: None or str | ||
| + | @keyword optimise_r2eff: Flag to specify if the read previous R2eff results should be optimised. For R1rho models where the error of R2eff values are determined by Monte-Carlo simulations, it can be valuable to make an initial R2eff run with a high number of Monte-Carlo simulations. Any subsequent model analysis can then be based on these R2eff values, without optimising the R2eff values. | ||
| + | @type optimise_r2eff: bool | ||
| + | @keyword insignificance: The R2eff/R1rho value in rad/s by which to judge insignificance. If the maximum difference between two points on all dispersion curves for a spin is less than this value, that spin will be deselected. This does not affect the 'No Rex' model. Set this value to 0.0 to use all data. The value will be passed on to the relax_disp.insignificance user function. | ||
| + | @type insignificance: float | ||
| + | @keyword numeric_only: The class of models to use in the model selection. The default of False allows all dispersion models to be used in the analysis (no exchange, the analytic models and the numeric models). The value of True will activate a pure numeric solution - the analytic models will be optimised, as they are very useful for replacing the grid search for the numeric models, but the final model selection will not include them. | ||
| + | @type numeric_only: bool | ||
| + | @keyword mc_sim_all_models: A flag which if True will cause Monte Carlo simulations to be performed for each individual model. Otherwise Monte Carlo simulations will be reserved for the final model. | ||
| + | @type mc_sim_all_models: bool | ||
| + | @keyword eliminate: A flag which if True will enable the elimination of failed models and failed Monte Carlo simulations through the eliminate user function. | ||
| + | @type eliminate: bool | ||
| + | @keyword set_grid_r20: A flag which if True will set the grid R20 values from the minimum R2eff values through the r20_from_min_r2eff user function. This will speed up the grid search with a factor GRID_INC^(Nr_spec_freq). For a CPMG experiment with two fields and standard GRID_INC=21, the speed-up is a factor 441. | ||
| + | @type set_grid_r20: bool | ||
| + | @keyword r1_fit: A flag which if True will activate R1 parameter fitting via relax_disp.r1_fit for the models that support it. If False, then the relax_disp.r1_fit user function will not be called. | ||
| + | """ | ||
| + | |||
| + | |||
| + | # Go | ||
| + | aa_relax_disp.Relax_disp(pipe_name=pipe_name, pipe_bundle=pipe_bundle, results_dir=result_dir_name, models=MODELS, grid_inc=GRID_INC, mc_sim_num=MC_NUM, exp_mc_sim_num=None, modsel=MODSEL, pre_run_dir=None, optimise_r2eff=False, insignificance=0.0, numeric_only=False, mc_sim_all_models=False, eliminate=True, set_grid_r20=set_grid_r20, r1_fit=r1_fit) | ||
| + | }} | ||
| + | |||
| + | === Run the same analysis in relax through terminal === | ||
| + | The analysis can be performed by | ||
| + | <source lang="bash"> | ||
| + | python 1_prepare_data.py | ||
| + | |||
| + | relax 2_load_data.py -t log.txt | ||
| + | # Or | ||
| + | mpirun -np 8 relax --multi='mpi4py' 2_load_data.py -t log.txt | ||
| + | </source> | ||
| + | |||
| + | == Inspect and make graphs == | ||
| + | |||
| + | * When this is done, quit relax | ||
| + | * Then to convert all xmgrace files to png | ||
| + | |||
| + | First make graphs | ||
| + | <source lang="bash"> | ||
| + | cd grace | ||
| + | ./grace2images.py -t EPS,PNG | ||
| + | |||
| + | cd .. | ||
| + | cd No_Rex/ | ||
| + | ./grace2images.py -t EPS,PNG | ||
| + | cat r1rho_prime.out | ||
| + | |||
| + | cd .. | ||
| + | cd M61 | ||
| + | ./grace2images.py -t PNG | ||
| + | cat r1rho_prime.out | ||
| + | cat kex.out | ||
| + | cat phi_ex.out | ||
| + | |||
| + | cat kex.out | grep "None 12" | ||
| + | cat phi_ex.out | grep "None 12" | ||
| + | </source> | ||
| + | |||
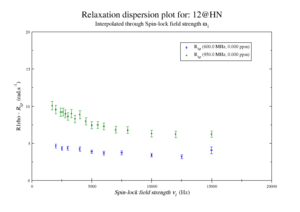
| + | This should give the images of the data | ||
| + | <gallery widths=300px heights=240px > | ||
| + | Disp 12 HN.png|Resi 12 at 600+950 MHz | ||
| + | </gallery> | ||
| + | |||
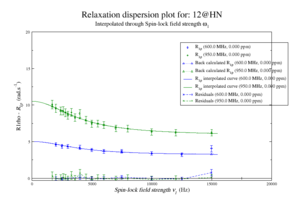
| + | For model [[M61]], spin 12 is: | ||
| + | <gallery widths=300px heights=240px > | ||
| + | File:Disp 12 HN M61.png|Resi 12 at 600+950 MHz for [[M61]] model. kex=25000, phi_ex=0.32 | ||
| + | </gallery> | ||
| + | |||
| + | == Further analysis steps == | ||
| + | |||
| + | Load the final state | ||
| + | |||
| + | * Inspect which residues should be included in analysis | ||
| + | <source lang="bash"> | ||
| + | # Now simulate that all spins are first deselected, and then selected. | ||
| + | deselect.all() | ||
| + | sel_ids = [ | ||
| + | ":12", | ||
| + | ":51", | ||
| + | ] | ||
| + | for sel_spin in sel_ids: | ||
| + | print("Selecting spin %s"%sel_spin) | ||
| + | select.spin(spin_id=sel_spin, change_all=False) | ||
| + | |||
| + | # Inspect which residues should be analysed together for a clustered/global fit. | ||
| + | cluster_ids = sel_ids | ||
| + | |||
| + | # Cluster spins | ||
| + | for curspin in cluster_ids: | ||
| + | print("Adding spin %s to cluster"%curspin) | ||
| + | relax_disp.cluster('model_cluster', curspin) | ||
| + | |||
| + | # Show the pipe | ||
| + | print("\nPrinting all the available pipes.") | ||
| + | pipe.display() | ||
| + | |||
| + | # Get the selected models | ||
| + | print("\nChecking which model is stored per spin.") | ||
| + | for curspin, mol_name, res_num, res_name, spin_id in spin_loop(full_info=True, return_id=True, skip_desel=False): | ||
| + | print("For spin_id '%s the model is '%s''"%(spin_id, curspin.model)) | ||
| + | |||
| + | # Copy pipe and switch to it. | ||
| + | pipe.copy(pipe_from="final - relax_disp", pipe_to="relax_disp_cluster", bundle_to="relax_disp_cluster") | ||
| + | pipe.switch(pipe_name="relax_disp_cluster") | ||
| + | pipe.display() | ||
| + | |||
| + | # Go again with clustered spins. | ||
| + | relax_disp.Relax_disp(pipe_name="relax_disp_cluster", pipe_bundle="relax_disp_cluster", results_dir=outdir+sep+"cluster", models=MODELS, grid_inc=GRID_INC, mc_sim_num=MC_NUM, exp_mc_sim_num=None, modsel=MODSEL, pre_run_dir=None, optimise_r2eff=False, insignificance=0.0, numeric_only=False, mc_sim_all_models=False, eliminate=True, set_grid_r20=set_grid_r20, r1_fit=r1_fit) | ||
| + | |||
| + | # Get the clustered fitted values | ||
| + | print("\nChecking which value is stored per spin.") | ||
| + | kex = None | ||
| + | for curspin, mol_name, res_num, res_name, spin_id in spin_loop(full_info=True, return_id=True, skip_desel=False): | ||
| + | if kex == None: | ||
| + | kex = curspin.kex | ||
| + | self.assertEqual(curspin.kex, kex) | ||
| + | print("For spin_id %s the kex is %.3f"%(spin_id, kex)) | ||
| + | </source> | ||
| + | |||
| + | * Run analysis again | ||
== See also == | == See also == | ||
[[Category:Tutorials]] | [[Category:Tutorials]] | ||
[[Category:Relaxation dispersion analysis]] | [[Category:Relaxation dispersion analysis]] | ||
Latest revision as of 10:00, 25 November 2015
Contents
Data background
This is data recorded at 600 and 950 MHz.
This should follow relax-4.0.1.Darwin.dmg installation on a Mac, only with GUI.
For each spectrometer frequency, the data is saved in np.arrays
- one for the residue number,
- one for the rates,
- one for the errorbars,
- one for the RF field strength.
They can be retrieved also with scipy's loadmat command.
The experiments are on-resonance R1ρ, and the rates are already corrected for the (small) offset effect, using the experimentally determined R1.
Specifically, the numpy shapes of the data is:
- For 600 MHz
- residues (1, 60)
- rates (60, 10)
- errorbars_rate (60, 10)
- RFfields (1, 10)
- For 950 Mhz
- residues (1, 61)
- rates (61, 19)
- errorbars_rate (61, 19)
- RFfields (1, 19)
For the 950 MHz, the 4000 Hz dispersion point is replicated.
An example of the data at the 2 fields is:
Create data files for relax
First prepare data, by running in Python:
python 1_prepare_data.py
The script is:
1_prepare_data.py script.Run analysis in relax GUI
- Start relax
- Then click: new analysis or ⌘ Cmd+n
- Then click icon for Relaxation dispersion → Next
- Just accept name for the pipe
- Open the interpreter: ⌘ Cmd+p
- Paste in
import os; os.chdir(os.getenv('HOME') + os.sep + 'Desktop' + os.sep + 'temp'); pwd()
- Then do
script(file='2_load_data.py')
- ( You can scroll through earlier commands with: ⌘ Cmd+↑ )
- Close the Grace
Results viewerwindow - Close the interpreter window
Now save the current state! This saved state can now be loaded just before an analysis, and contain all setup and data.
- File → Save as (Shift+Ctrl+S) → Save as
before_analysis.bz2
Now Quit relax, and start relax again.
Go to: File → Open relax state (Ctrl+O) → Locate before_analysis.bz2
In the GUI, select:
- For models, only select:
No RexandM61 - R1 parameter optimisation to False, as this on-resonance data does not use R1.
- The number of Monte-Carlo simulations to
3. (The minimum number before relax refuse to run). - Let other things be standard, and click execute
| Note The number of Monte-Carlo simulations is set to 3. This means that relax will do the analysis, and the fitted parameters will be correct, but the standard error of the parameters will be wrong. |
The reason for this is, that the data "naturally"? for some spins contains measurements which are "doubtful".
When relax is performing Monte-Carlo simulations, all the data is first copied x times to the number of Monte-Carlo simulations, and each R2,eff point on the graph
is modified randomly by a gaussian noise with a width described by the associated errors.
For spins which contains measurements which are "doubtful", relax will be spending "very long time" trying to fit a meaningful model.
This "time" is poorly spend on "bad data".
Rather, one should first try to
- analyse the data quickly
- make the graphs
- examine which spins should be deselected
- and then rerun the analysis with a higher number of monte-carlo simulations
After this comes the part with global fit/clustered analysis.
- analyse the data again
- Select which spins should be included in a global fit/clustered analysis
- and then rerun with analysis
The script
2_load_data.py script.Run the same analysis in relax through terminal
The analysis can be performed by
python 1_prepare_data.py
relax 2_load_data.py -t log.txt
# Or
mpirun -np 8 relax --multi='mpi4py' 2_load_data.py -t log.txt
Inspect and make graphs
- When this is done, quit relax
- Then to convert all xmgrace files to png
First make graphs
cd grace
./grace2images.py -t EPS,PNG
cd ..
cd No_Rex/
./grace2images.py -t EPS,PNG
cat r1rho_prime.out
cd ..
cd M61
./grace2images.py -t PNG
cat r1rho_prime.out
cat kex.out
cat phi_ex.out
cat kex.out | grep "None 12"
cat phi_ex.out | grep "None 12"
This should give the images of the data
For model M61, spin 12 is:
Resi 12 at 600+950 MHz for M61 model. kex=25000, phi_ex=0.32
Further analysis steps
Load the final state
- Inspect which residues should be included in analysis
# Now simulate that all spins are first deselected, and then selected.
deselect.all()
sel_ids = [
":12",
":51",
]
for sel_spin in sel_ids:
print("Selecting spin %s"%sel_spin)
select.spin(spin_id=sel_spin, change_all=False)
# Inspect which residues should be analysed together for a clustered/global fit.
cluster_ids = sel_ids
# Cluster spins
for curspin in cluster_ids:
print("Adding spin %s to cluster"%curspin)
relax_disp.cluster('model_cluster', curspin)
# Show the pipe
print("\nPrinting all the available pipes.")
pipe.display()
# Get the selected models
print("\nChecking which model is stored per spin.")
for curspin, mol_name, res_num, res_name, spin_id in spin_loop(full_info=True, return_id=True, skip_desel=False):
print("For spin_id '%s the model is '%s''"%(spin_id, curspin.model))
# Copy pipe and switch to it.
pipe.copy(pipe_from="final - relax_disp", pipe_to="relax_disp_cluster", bundle_to="relax_disp_cluster")
pipe.switch(pipe_name="relax_disp_cluster")
pipe.display()
# Go again with clustered spins.
relax_disp.Relax_disp(pipe_name="relax_disp_cluster", pipe_bundle="relax_disp_cluster", results_dir=outdir+sep+"cluster", models=MODELS, grid_inc=GRID_INC, mc_sim_num=MC_NUM, exp_mc_sim_num=None, modsel=MODSEL, pre_run_dir=None, optimise_r2eff=False, insignificance=0.0, numeric_only=False, mc_sim_all_models=False, eliminate=True, set_grid_r20=set_grid_r20, r1_fit=r1_fit)
# Get the clustered fitted values
print("\nChecking which value is stored per spin.")
kex = None
for curspin, mol_name, res_num, res_name, spin_id in spin_loop(full_info=True, return_id=True, skip_desel=False):
if kex == None:
kex = curspin.kex
self.assertEqual(curspin.kex, kex)
print("For spin_id %s the kex is %.3f"%(spin_id, kex))
- Run analysis again