Difference between revisions of "Relax disp.spin lock offset+field"
| Line 20: | Line 20: | ||
Palmer, A.G. & Massi, F. (2006). Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem. Rev. 106, 1700-1719 [http://dx.doi.org/10.1021/cr04042875 DOI] | Palmer, A.G. & Massi, F. (2006). Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem. Rev. 106, 1700-1719 [http://dx.doi.org/10.1021/cr04042875 DOI] | ||
| − | [[File:Fig1 Palmer Massi 2006.png|thumb|center|upright= | + | [[File:Fig1 Palmer Massi 2006.png|thumb|center|upright=3|Try to reproduce Figure 1.]] |
Figure produced with script [[Relax_disp.spin_lock_offset%2Bfield_figure | found here. ]] | Figure produced with script [[Relax_disp.spin_lock_offset%2Bfield_figure | found here. ]] | ||
| Line 32: | Line 32: | ||
<source lang="python"> | <source lang="python"> | ||
offsets[ei][si][mi][oi] = frequency_to_rad_per_s(frq=cdp.spin_lock_offset[id], B0=frq, isotope=spin.isotope) | offsets[ei][si][mi][oi] = frequency_to_rad_per_s(frq=cdp.spin_lock_offset[id], B0=frq, isotope=spin.isotope) | ||
| − | |||
| − | |||
</source> | </source> | ||
=== spin lock field === | === spin lock field === | ||
The spin lock field strength is noted '''$\nu_1$'''.<br> | The spin lock field strength is noted '''$\nu_1$'''.<br> | ||
| − | The [[wikipedia:Chemical_shift | Chemical Shifts]] $\delta$ in ppm for nuclei of interest (ex. $^{15}$N and which have been loaded in with relax function [http://www.nmr-relax.com/manual/chemical_shift_read.html chemical_shift_read] from a [http://www.nmr-relax.com/manual/spectrum_read_intensities.html peak list formatted file]) is first converted to | + | The [[wikipedia:Chemical_shift | Chemical Shifts]] $\delta$ in ppm for nuclei of interest (ex. $^{15}$N and which have been loaded in with relax function [http://www.nmr-relax.com/manual/chemical_shift_read.html chemical_shift_read] from a [http://www.nmr-relax.com/manual/spectrum_read_intensities.html peak list formatted file]) is first converted to to the rad/s with the following function calls. |
| + | <source lang="python"> | ||
| + | shifts[ei][si][mi] = frequency_to_rad_per_s(frq=shift, B0=frq, isotope=spin.isotope) | ||
| + | </source> | ||
=== The trouble === | === The trouble === | ||
Revision as of 11:49, 16 March 2014
Contents
Setting up relax_disp.spin_lock_offset and relax_disp.spin_lock_field
Refer to the manual for parameter explanation
This page is a little help to understand how to use the functions:
- relax_disp.spin_lock_offset()
- relax_disp.spin_lock_field()
spin lock offset
Manual on relax_disp.spin_lock_offset
The relax function relax_disp.spin_lock_offset() requires the values to be provided in ppm.
relax_disp.spin_lock_offset(spectrum_id=None, offset=None)
spin lock field
Manual on relax_disp.spin_lock_field
The relax function relax_disp.spin_lock_field() requires the values to be provided in Hz.
relax_disp.spin_lock_field(spectrum_id=None, field=None)
Literature comments
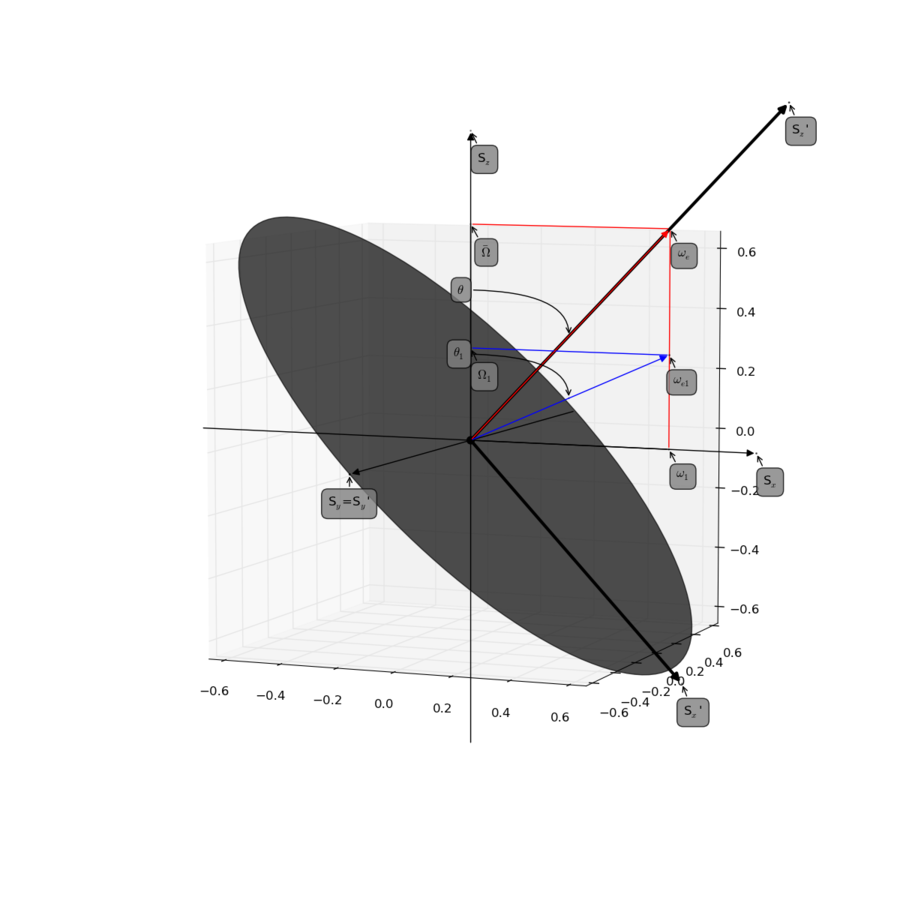
See Figure 1 and 10 in the reference.
Palmer, A.G. & Massi, F. (2006). Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem. Rev. 106, 1700-1719 DOI
Figure produced with script found here.
spin lock offset
The offset is in the literature noted as $\Omega$, where $\Omega$ is the $^{15}$N resonance offset from the spin-lock carrier.
In the literature, the values are often stated as "offset", "carrier offset", "offset of the spin-lock pulse" with values given in Hz, and have values from 0-500 to 10-20.000 Hz.
Relax needs input in ppm, and converts to the rad/s, with the following function calls.
offsets[ei][si][mi][oi] = frequency_to_rad_per_s(frq=cdp.spin_lock_offset[id], B0=frq, isotope=spin.isotope)
spin lock field
The spin lock field strength is noted $\nu_1$.
The Chemical Shifts $\delta$ in ppm for nuclei of interest (ex. $^{15}$N and which have been loaded in with relax function chemical_shift_read from a peak list formatted file) is first converted to to the rad/s with the following function calls.
shifts[ei][si][mi] = frequency_to_rad_per_s(frq=shift, B0=frq, isotope=spin.isotope)
The trouble
The trouble is.
Does the Hz frequency refers to RF fields applied at the 1H Larmor frequency or 15N frequency?
At page 1708 is states that w_1S = w_1 and w_eS = w_e. And in pulse sequence it states that:
Code reference calculations in relax
The code which is called resides in:
lib/nmr.py
frequency_to_rad_per_s(frq=None, B0=None, isotope=None):
specific_analyses/relax_disp/disp_data.py
return_offset_data(spins=None, spin_ids=None, field_count=None, fields=None):
Spectrometer notes
Varian / VnmrJ
In some pulse sequences, the following is seen:
'trim' is a basic timeunit and the total spinlock time is calculated as 2.0*ncyc*trim
b1 = getval("b1"), /* spin-lock field, Hz! */
deltadof2 = getval("deltadof2"), /* offset for N15 spinlock */