Welcome to the relax Wiki.
| The community-run support site for relax, a program for the study of molecular dynamics using experimental NMR data.
|
User contributions - How to edit pages at the wiki
Please read the guidelines here.
What's new?
|
|
Did you know...
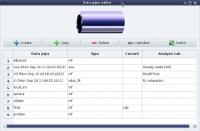
Custom peak list
protein 3 R 3 N 3.642647e+06 6.056554e+06 3.753433e+06 ...
protein 5 E 5 N 1.609356e+06 2.927111e+06 1.726433e+06 ...
protein 6 V 6 N 1.697771e+06 3.015788e+06 1.771777e+06 ...
protein 7 N 7 N 1.535896e+06 3.005234e+06 1.683477e+06 ...
protein 8 I 8 N 1.332059e+06 2.611723e+06 1.519182e+06 ...
protein 9 V 9 N 1.532644e+06 2.834876e+06 1.597882e+06 ...
protein 11 N 11 N 1.219399e+06 2.479556e+06 1.289455e+06 ...
protein 15 D 15 N 2.808704e+06 4.812759e+06 2.935094e+06 ...
protein 16 Q 16 N 3.410860e+06 5.823456e+06 3.427248e+06 ...
protein 22 E 22 N 3.681818e+06 6.378086e+06 3.663392e+06 ...
...
Sequence read
<source lang="python">
sequence.read(file='Custom_model.txt', dir=None, spin_id_col=None, mol_name_col=1, res_num_col=2, res_name_col=3, spin_num_col=4, ..→
|
|
|
Random screenshots
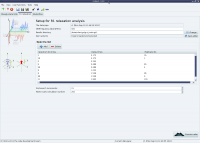
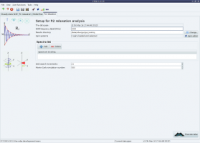
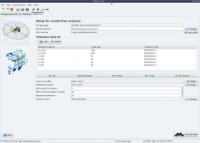
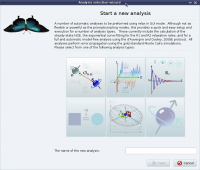 The analysis selection wizard  Steady-state NOE analysis
|
|