Welcome to the relax Wiki.
| The community-run support site for relax, a program for the study of molecular dynamics using experimental NMR data.
|
User contributions - How to edit pages at the wiki
Please read the guidelines here.
What's new?
|
|
Did you know...
Coding editor
A lightweight editor, could be pyscripter.
This helps with syntax high-lighting and includes the interpreter.
Install from: https://code.google.com/p/pyscripter/
To let pyscripter find winpython, you need to register it in the windows registry.
Advanced->Register distribution->Yes
Setup
Indention
Remember to check that indention is set to equal 4 spaces " ", under Tools/Options/Editor Options
Trailing spaces
Some parts of relax requires trailing whitespace.
Tools/Options/Editor Options/Options/ Untick "Trim Trailing spaces"
Run engine
Also set: Run/Python Engine/Remote (Tk) (Matplotlib uses the Tk)
Test
Test that everything is working, for example by this script, and hit the green play button.
from pylab import *
a ..→
|
|
|
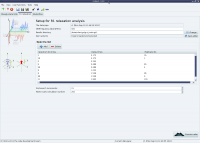
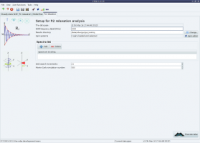
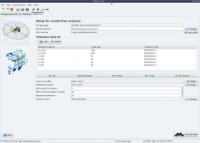
Random screenshots
 The analysis selection wizard  Steady-state NOE analysis
|
|