Welcome to the relax Wiki.
| The community-run support site for relax, a program for the study of molecular dynamics using experimental NMR data.
|
User contributions - How to edit pages at the wiki
Please read the guidelines here.
What's new?
|
|
Did you know...
Open MPI is a Message Passing Interface (MPI) used in relax for parallalised calculations.
mpi4py OpenMPI
This package provides Python bindings for the Message Passing Interface (MPI) standard. It is implemented on top of the MPI-1/2/3 specification and exposes an API which grounds on the standard MPI-2 C++ bindings.
relax manual on Multi processor usage
If you have OpenMPI and mpi4py installed, then you have access to Gary Thompson's multi-processor framework for MPI parallelisation.
Gary has achieved near perfect scaling efficiency:
https://www.nmr-relax.com/mail.gna.org/public/relax-devel/2007-05/msg00000.html
Dependencies
- Python 2.4 to 2.7 or 3.0 to 3.4, or a recent PyPy release.
- A functional MPI 1.x/2.x/3.x implementation like MPICH or Open MPI built with shared/dynamic ..→
|
|
|
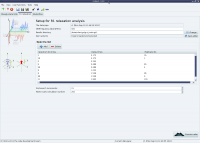
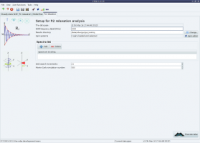
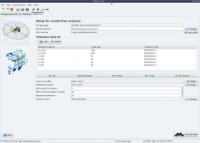
Random screenshots
 The analysis selection wizard  Steady-state NOE analysis
|
|